How to do NGS 50% faster
The Eurofins Genomics NGS capabilities
The Eurofins Genomics NGS facility welcomed two new members to the NGS platform fleet: The Nanopore GridION and another Illumina NovaSeq 6000 (image 1). This addition increases our sequencing capacities and flexibility even further, but almost more importantly, it affects another crucial factor: TIME!
The need to quickly execute and realise a project, be it in research or production, is now more important than ever. We are living in a time of “overnight” and “next day” delivery.
Similar to setting the TAT industry standard for Sanger sequencing to overnight in 2007 (the time for sample sequencing and delivery of results is called turnaround time), we have now made it our mission to provide the fastest TAT possible for NGS.

Image 1: Nanopore GridION and Illumina NovaSeq 6000.
More than NGS platforms: The pivotal factors for a fast NGS TAT
Several factors are key for a fast TAT. Our high NGS capacities form the basis, but it is not all about capacity. It is about how capacity is implemented. With years of experience and expertise, we quickly applied our NGS capacities to gradually shorten the TAT for our NGS solutions:

An additional key factor for our fast TATs is the Lean management of the entire NGS process, from sample reception, processing and sequencing, to BioIT and data delivery. Just as important is the high degree of process automation and the utilisation of an NGS laboratory information management system (NGS-LIMS).

Lean management
Our laboratory processes are constantly Lean-optimised by our skilled Lean managers (image 2). We also adhere to the Six Sigma principles. In-depth analysis, extensive planning and thorough testing enabled us to provide a much more efficient and streamlined structure of our NGS processes.
Image 2: Drafting stage of the Lean optimisation of an entire NGS process from sample reception to data delivery.
Automation of the lab process
Sample preparation in our NGS labs is highly automated and utilises liquid handling robots (image 3). This ensures a high degree of consistency across samples and biological and technical replicas.
Meticulous maintenance of our liquid handling robots ensures consistent, high-quality performance during the sample preparation.

Image 3: Example of the liquid handling robots utilised in the NGS lab for sample preparation.
The NGS platforms
When it comes to DNA and RNA high-throughput sequencing, Eurofins Genomics uses the most reliable, proven and market-leading NGS platforms.
The Illumina NGS platforms are the market-leading NGS devices and excel in output, accuracy and quality of sequencing. They are based on the sequencing-by-synthesis technology. The proven and well-established platforms NovaSeq 6000 and MiSeq allow us to provide NGS data from 13 Gb to 6,000 Gb per run (image 4).
The Nanopore GridION from Oxford Nanopore Technologies stands out with exceptional read lengths of more than 4,000,000 bp (4 Mb). It is based on the latest nanopore sequencing technology and provides NGS data up to 50 Gb per run (image 4).
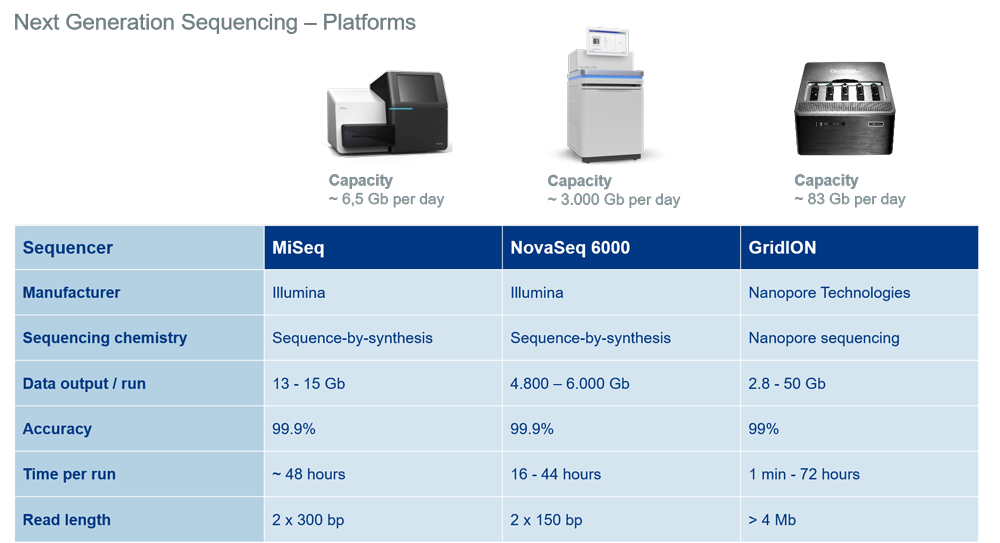
Image 4: Overview of NGS platforms and their specifications at Eurofins Genomics.
All procedures in the NGS lab are supported by our NGS laboratory information management system (NGS-LIMS). It is an automated system that monitors all individual steps of the preparation and sequencing process. The NGS-LIMS provides regular updates on the processing status of the samples and displays it in the Eurofins Genomics web shop (image 5). Our customers can track the status of their samples anytime, so they can adjust their planning and follow-up work accordingly.
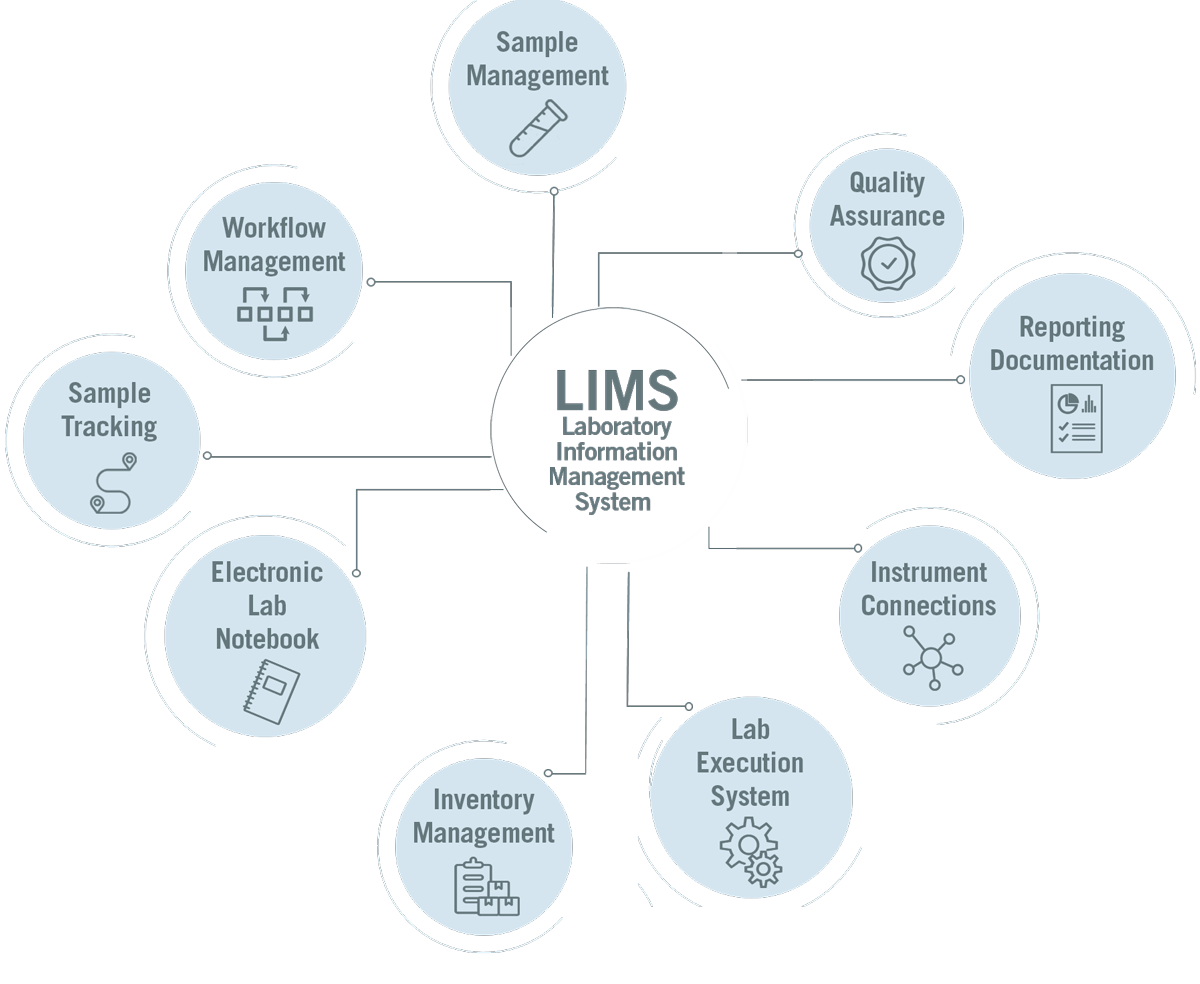
Image 5: Depiction of the NGS-LIMS functionalities.
Sequencing on NGS platforms is only as good as the DNA and RNA extraction
Nucleic acid extraction can be tricky, especially when you lack experience in DNA and RNA extraction. The Eurofins Genomics experts have long-standing experience in DNA and RNA extraction that is used daily in labs. We have a proven track record of cooperation with many large-scale research projects where we carried out DNA/RNA extraction and high-throughput sequencing.
Our customers often outsource the DNA or RNA extraction step of their project to us. We extract DNA and RNA from all types of samples, including animal tissue and samples, cells and blood, bacterial cell pellets, plant tissue and environmental samples (image 6).
Image 6: Overview of samples for DNA and RNA extraction.
The utilised extraction method is specifically selected according to the amount of DNA or RNA that is needed in the downstream process and according to the amount of sample material available.
DNA and RNA are extracted in automated systems by using established bead-based kits or column-based kits.
Bead-based extraction is done on platforms such as the KingFisherTM Flex System, Chemagic™ instruments and QIASymphony® (image 7).
The automated DNA and RNA extraction using column based methods is done on the QIAGEN QIACube®. It is used for automated simultaneous processing of up to 12 samples per run (image 8).

Image 8: QIACube® for DNA and RNA extraction using column based methods.
Depending on the project, we also provide manual DNA and RNA extraction from tissues and cells by using established column-based kits (e. g. Qiagen RNEasy® Mini).
DNA and RNA extraction involves many steps in the lab, especially when manual extraction is required. A careful and thorough Lean optimisation of the lab and extraction process leads to constant optimisation and, eventually, streamlined lab processes in our NGS facility.
Lean optimisation of the workspace and benches is extremely beneficial for the process. Here, unnecessary equipment, solutions and consumables are removed to clear benches. The devices are grouped in sequence with the process to avoid long distances (image 9).


Image 9: Example of the optimisation of the lab space and waste elimination.
But doesn’t fast processing and fast TATs mean that quality has to be sacrificed? How does Eurofins Genomics guarantee high data quality?
How to make the most of NGS platforms: Quality control
The quality and quantity of each DNA and RNA sample is determined shortly after arrival at our NGS facility and prior to sequencing. For input sample QC, intermediate library QC and final library QC, we use the gold standards of methods: Nanodrop, Qubit Fluorometer and Agilent 2100 Bioanalyzer (image 10).
The Nanodrop device operates based on spectrophotometric quantification and measures DNA/RNA concentration and purity.
The Qubit Fluorometer is based on fluorometric quantification and measures DNA/RNA concentration and purity with very high accuracy, however, it gives no information on the DNA/RNA integrity.
The Agilent 2100 Bioanalyzer, based on capillary electrophoresis, is an automated tool for assessing DNA and RNA quantity, fragment size distribution and degradation degree. It is also used to monitor the size distribution after fragmentation and adapter ligation.

Image 10: Utilised devices to measure and assess the quality and quantity of samples: a) Nanodrop, b) Qubit Fluorometer, and c) Agilent 2100 Bioanalyzer.
The NGS workflow for our NGS platforms
Customers send their samples to the Eurofins NGS facility, where we provide high-throughput sequencing for requests ranging from one to thousands of samples according to customer needs.
According to the type of samples and customer requirements, we use automated or manual DNA/RNA extraction based on bead or column kits. Following this, the DNA/RNA quality and quantities are checked.
For exome sequencing for instance, the DNA library is prepared using an optimised protocol and the latest enrichment technologies (Agilent SureSelect panel) for highest coverage, even of regions that are hard to capture.
The Illumina technology allows for the pooling of numerous samples, called multiplexing, for processing in parallel. High-throughput sequencing of the pooled samples is performed on the NovaSeq 6000 platform, which can generate up to 6,000 Gb of sequencing data.
In order to prevent sample misassignment in multiplexed NGS by index hopping, we utilise UDI’s (unique dual indexing) for library preparation. This represents the most advanced and reliable indexing standard for NGS.
The sequencing data produced is then analysed in our BioIT department via a bioinformatics pipeline to filter exome variants such as SNPs, then the data is annotated and interpreted. Variant discovery includes the mapping of the data against a reference genome, gene IDs and effects on protein level (affected amino acids), and more (image 11).
The sequencing data and BioIT analyses are then provided to our customers. However, due to increasing digitalisation, we put particular emphasis on data security and measures to securely deliver customer data.
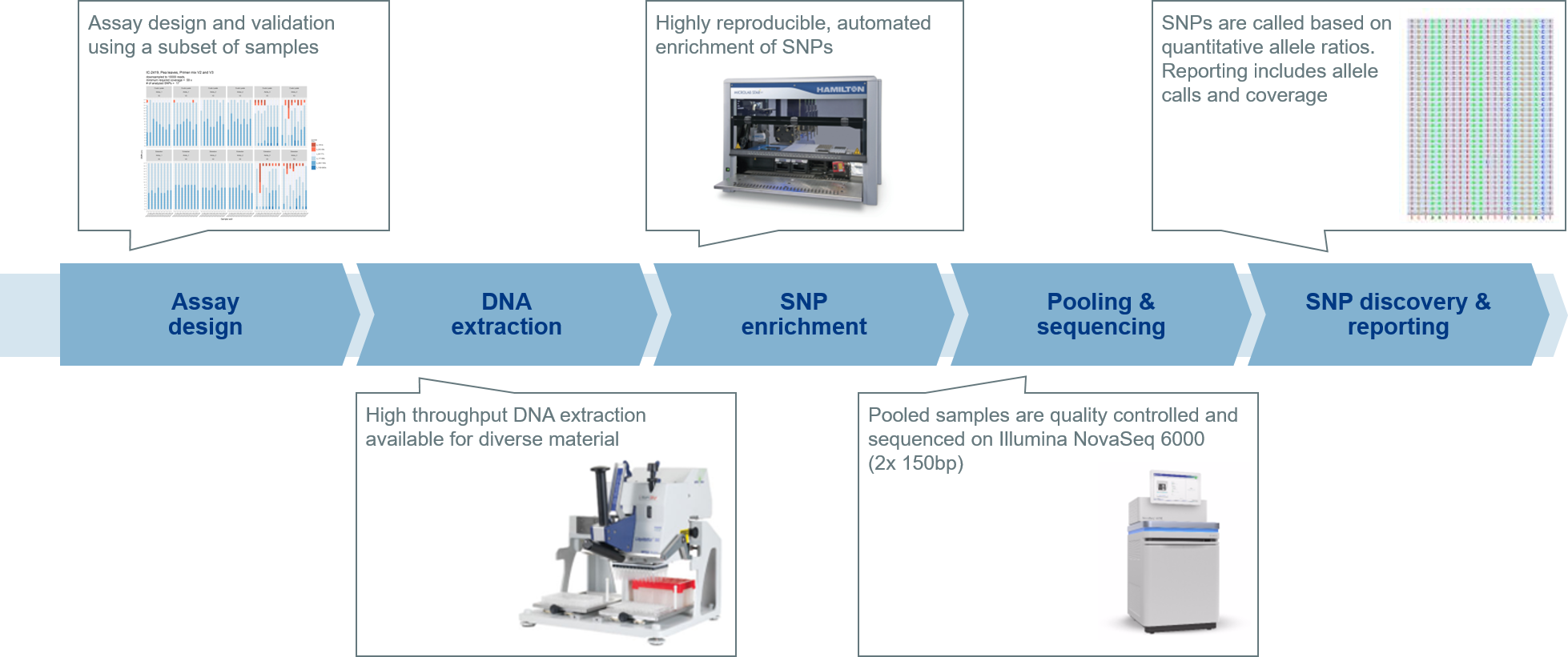
Image 11: Depiction of the NGS workflow using the example of single nucleotide polymorphisms (SNPs) detection and discovery.
Data protection for NGS
For confidential delivery of customer data, we utilise secure FTP servers (SFTP) and the secure Eurofins Genomics webshop. Only the project owner is granted access to the data via these secure access points (image 12). Data will never be provided via email or external file hosting.
We also operate under the General Data Protection Regulation (GDPR).
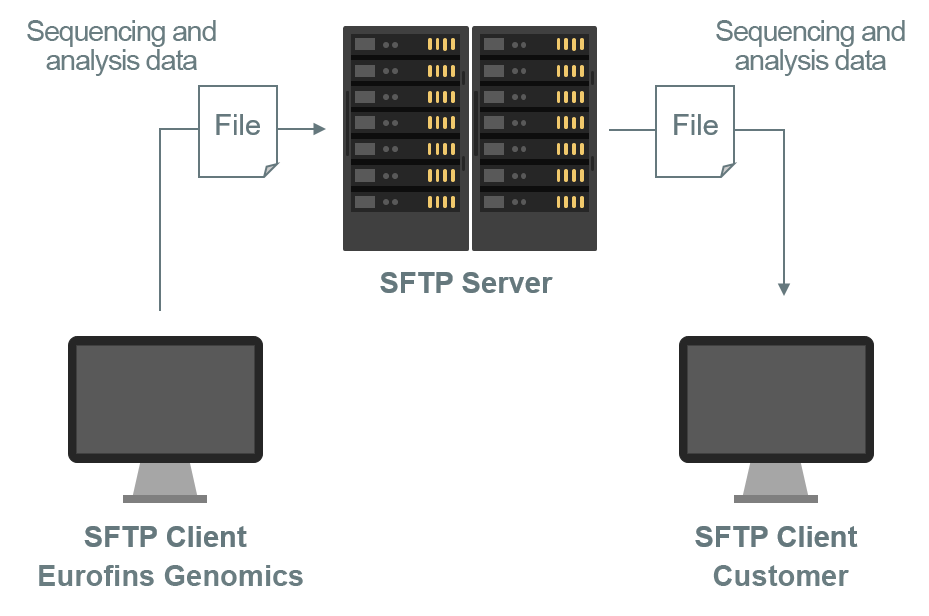
Image 12: Depiction of secure and confidential data delivery to customers.
NGS solutions
Our customers gain access to our range of NGS platforms, technical knowledge and expertise in all aspects of the NGS workflow, from optimised library preparation to professional BioIT analysis of all sequencing data.
Our NGS platforms along with our team of dedicated lab technicians and project managers are the basis for our NGS solutions. These range from RNA sequencing, microbiome/metagenome, exome and genome sequencing as predesigned, to modular and more cost-efficient solutions.
Are you not sure which NGS solution you need for your project? We designed a decision tree to find the NGS solutions that are most suitable for the individual project requirements of our customers.
NGS platforms: The technologies
Illumina’s sequencing-by-synthesis (SBS) technology is based on sequencing libraries. Here, the DNA extracted from a sample is randomly fragmented into shorter DNA pieces that are around 200 bases long. The entirety of all of these DNA fragments is called a library. It could be described as the DNA fragments representing the books in a library and the nucleotides representing the text/characters in the books.

Adaptors are then added to the ends of the DNA fragments. Following that, the double stranded DNA (dsDNA) is applied to the matrix, called flow cell, in the Illumina device (image 13). On these flow cells, the sequencing reaction takes place. Here, the dsDNA fragments are denatured into single stranded DNA (ssDNA) fragments that bind via their adaptors to oligonucleotide anchors that are fixed to the surface of the flow cell. The binding occurs via complementary sequences and, therefore, the capturing of the DNA sample on the flow cell.

Image13: Illumina NovaSeq 6000 flow cell.
Adaptors are then added to the ends of the DNA fragments. Following that, the double stranded DNA (dsDNA) is applied to the matrix, called flow cell, in the Illumina device (image 13). On these flow cells, the sequencing reaction takes place. Here, the dsDNA fragments are denatured into single stranded DNA (ssDNA) fragments that bind via their adaptors to oligonucleotide anchors that are fixed to the surface of the flow cell. The binding occurs via complementary sequences and, therefore, the capturing of the DNA sample on the flow cell.
Once the DNA fragments bind to the oligo anchors, polymerase, nucleotides, etc. are added and, according to the bound DNA fragment, a new strand is synthesised that is physically linked to the flow cell (image 14, steps 1-2).
The resulting dsDNA is denatured and the newly synthesised ssDNA fragments function as templates for amplification to form clusters of double stranded DNA (dsDNA) fragments. During this cluster generation process, DNA polymerases fill-in the ssDNA templates to generate dsDNA. As the ssDNA templates form bridge-like structures, this step is called bridge amplification. In fact, thousands of copies of each DNA template are created in tight clusters on the flow cell surface (image 14, steps 3-7).
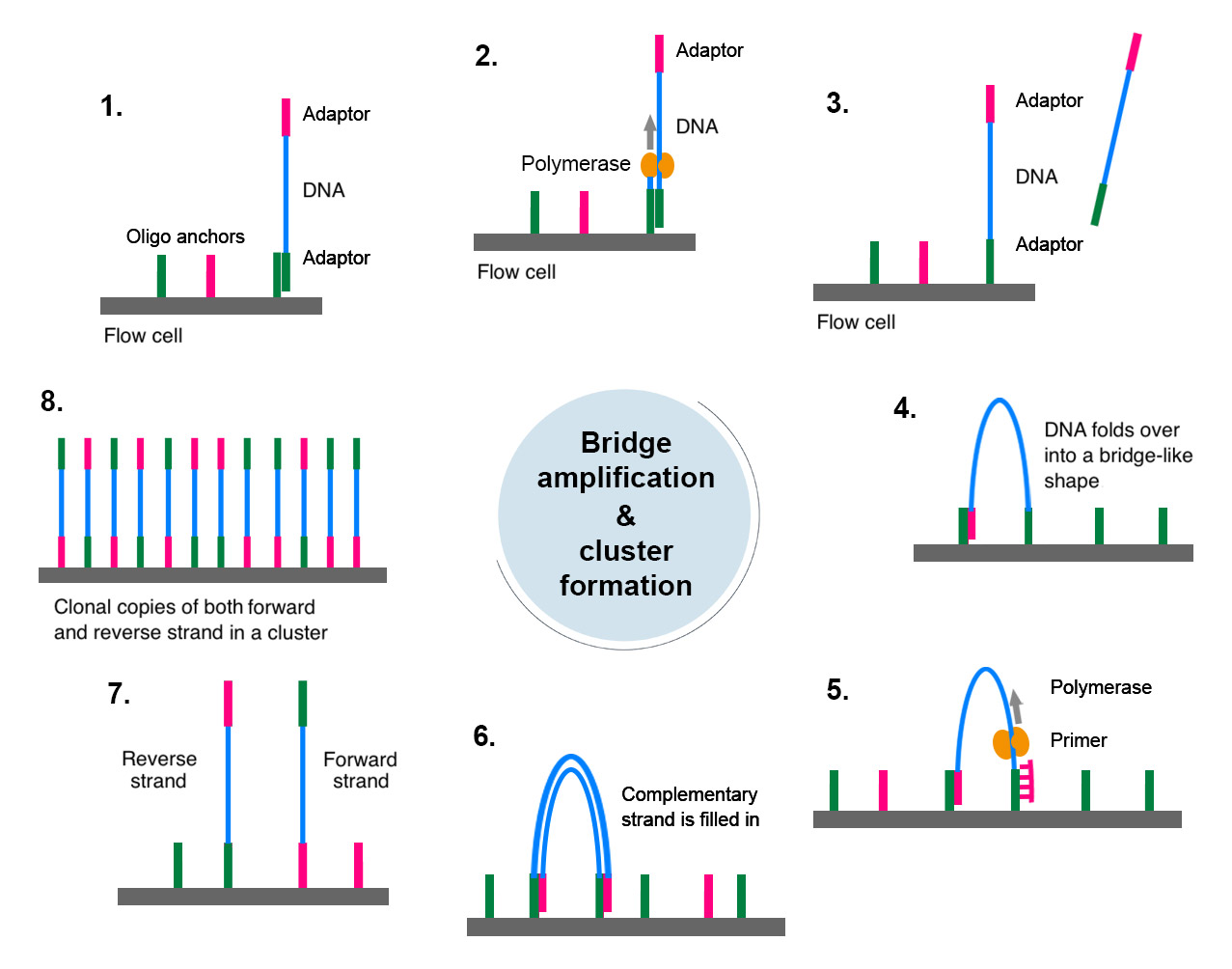
Image 14: Principle behind the bridge amplification and cluster generation steps of the sequencing-by-synthesis technology: 1) the ssDNA fragments bind to the oligonucleotide anchors on the flow cell via complementary sequences, 2) a DNA polymerase synthesises the complementary strand that is anchored to the flow cell, 3) the original template is washed away (after denaturation), 4) the new ssDNA bends and binds to another oligo anchor nearby to form a bridge-like structure, 5) a DNA polymerase incorporates nucleotides according to the template, which is called bridge amplification, 6) upon the fill-in reaction, the dsDNA is denatured into two ssDNA that straighten up, 7) both ssDNAs function as templates for another round of amplification resulting in a cluster of forward and reverse ssDNA clones.
Annotation
In recent years, Illumina changed the fluorescence dye system from four colours, where each nucleotide has a specific colour, to two. Now, the four bases are represented by red and green: C and T are represented by red and green respectively, A by a mix of red and green (both are visible in images) and G has no colour. The NovaSeq 6000 utilises the two-colour system that has been refined to the point where the data quality rivals the quality from the earlier four-colour system.
For the sequencing reaction, the dsDNA is denatured into ssDNA again and the reverse strands are cleaved and washed off. Following this, primers attach to the forward strands (ssDNA templates) and fluorescent dye-labelled nucleotides (A, T, G, C) are added.
Additionally, the fluorescent dyes attached to the nucleotides act as reversible terminators, also called blocking groups, and ensure that only one nucleotide is added to the synthesised strand at a time. Significantly, the fluorescent dye on each of the four nucleotides emits fluorescence light of a characteristic wavelength when incorporated into the DNA. These fluorescent light signals are detected by a camera and the data is used to determine the sequence of the DNA fragment. After every incorporation of a nucleotide and detection of the corresponding fluorescence signal, the blocking group and the fluorescence dye are chemically removed (deblocking step), allowing for another nucleotide to be incorporated. This process is then repeated until the entire DNA template is sequenced-by-synthesis (image 15).
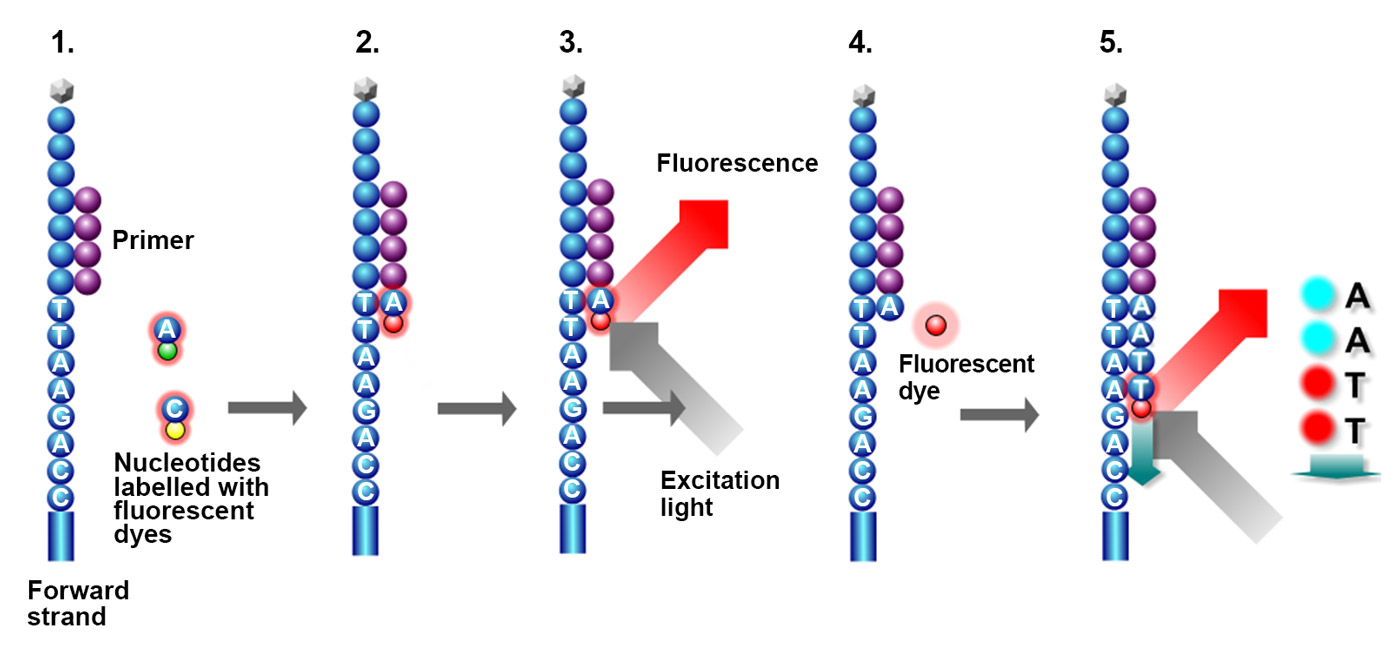
Image 15: Principle of sequencing-by-synthesis: 1) primer binds to complementary sequence of the ssDNA template, 2) fluorescent dye-labelled nucleotides are incorporated by DNA polymerase (not shown) according to the template, 3) fluorescent dye is excited and emits a fluorescence signal that is detected, 4) fluorescence dye is removed, 5) the next nucleotide is incorporated and the emitted fluorescence signal is detected.
The nanopore technology in the Nanopore GridION (the latest addition to our NGS platforms) is based on the detection of changes in electrical conductivity that arise when DNA strands are run through pores the size of nanometres.
Nanopores (~1.8 nm) are embedded in a lipid membrane and a currency is applied. The lipid membrane is electrically resistant and, thus, the ionic current has to pass through the nanopore.
As DNA is negatively charged, it moves through the nanopore, where a detector, the nanopore reader, is positioned. The detector measures the disruption of electrical current in the nanopore when a molecule such as DNA comes into contact with the nanopore.
During the preparation step, a motor protein is attached to the dsDNA and functions as a molecular velocity control for the passing of the DNA through the nanopore. The motor protein also functions as a helicase and separates the two strands from each other. The motor protein then feeds one ssDNA through the nanopore, where its sequence is determined by the nanopore reader. The combination of data from thousands of pores leads to a highly accurate sequencing result (base call) (images 16 and 17).
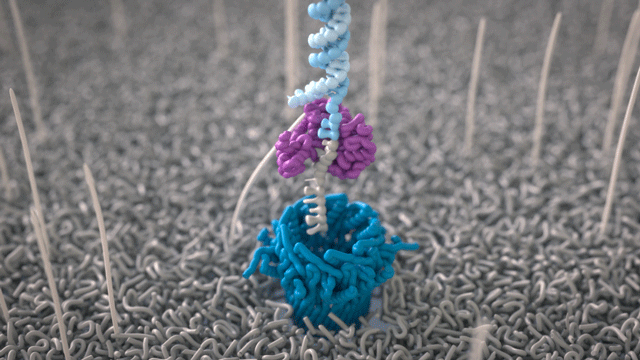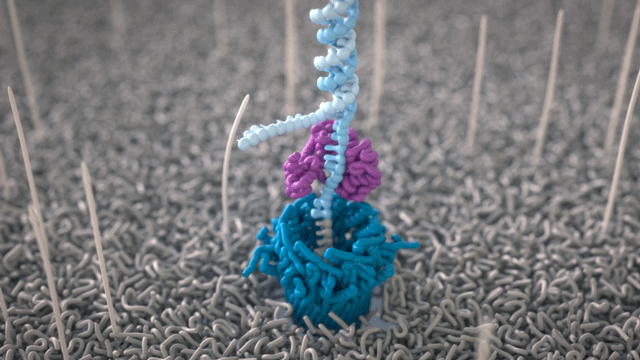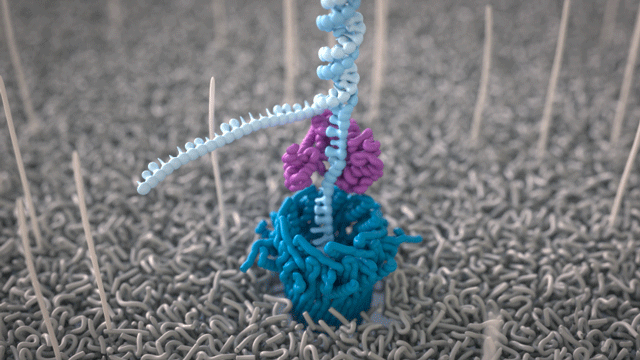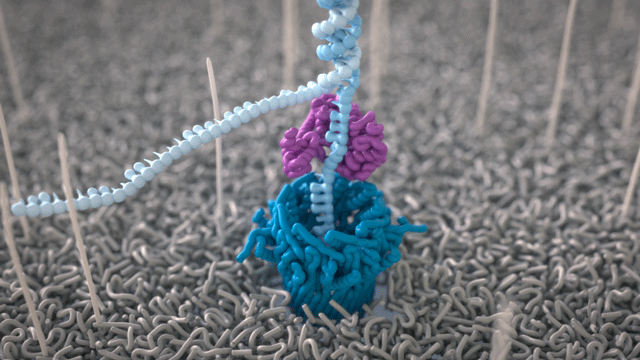
Image 16: Principle of the nanopore technology for DNA and RNA sequencing – part 1.
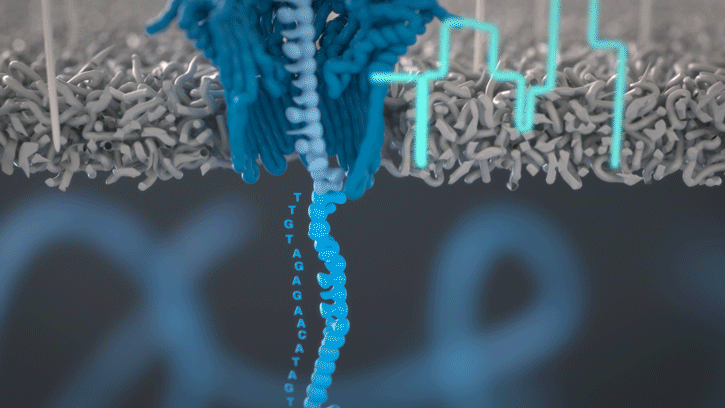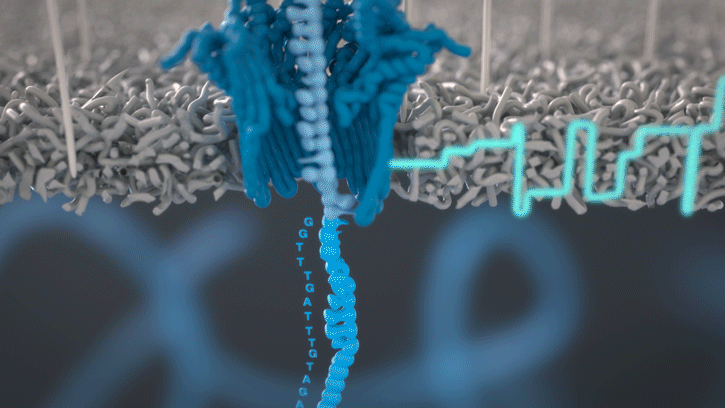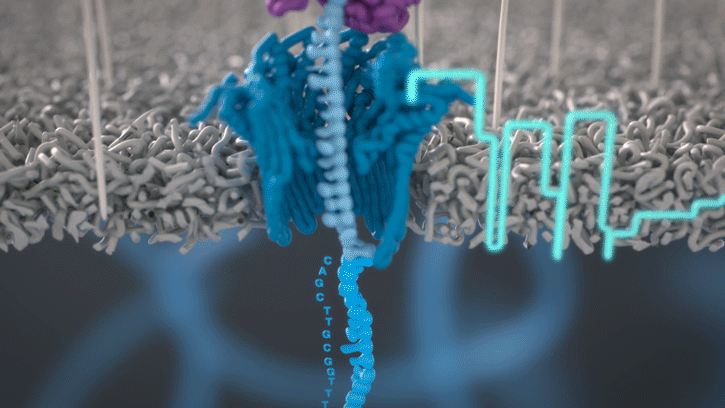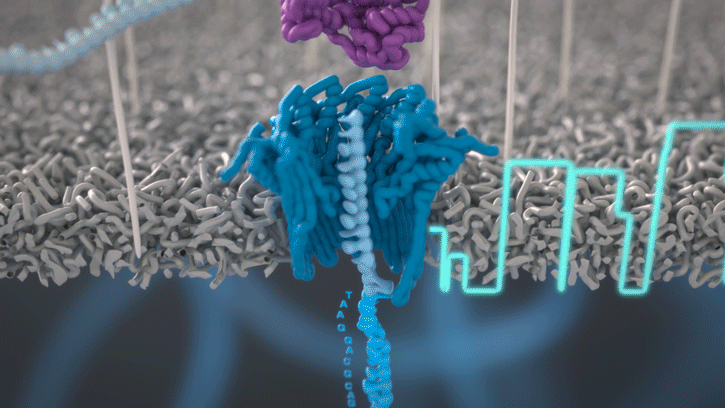
Image 17: Principle of the nanopore technology for DNA and RNA sequencing – part 2.
Eurofins Genomics: A trusted genomics solution provider
Eurofins Genomics is a trusted sequencing solutions provider with a certified product portfolio. The combination of in-depth knowledge in genomics and technical capabilities with innovative ideas has allowed us to claim and maintain the top spot as the leading sequencing services provider in Europe.
The trust placed in us is also reflected by our many collaborations with renowned small-, medium- and large-scale organisations and research institutes. Here, we provide leading DNA and RNA sequencing and analysing expertise combined with the state-of-the-art technologies to execute it.
Lately, we successfully cooperated with the IMI2 INNODIA consortium where we supported research on type 1 diabetes with large-scale RNA sequencing, and the SIMPLER research infrastructure, where we processed 38,000 samples and ran 40,500 samples on genome-wide association study microarrays.
With our high NGS capacities, Eurofins Genomics has also sequenced more than 100,000 SARS-CoV-2 samples from 23 European countries in just three months.
NGS platforms and our certificates and accreditations: Quality you can trust
Our quality standards are approved by several external certifications.

ISO 9001
Eurofins Genomics is certified according to ISO 9001. Our quality management (QM) system is highly developed and we are experienced in attending customer needs and expectations.
ISO 17025
The German authority DAkkS confirmed our high competence in sequencing analyses. The German accreditation is internationally accepted by the International Laboratory Accreditation Cooperation (ILAC).


ISO 13485
We are certified for the development of in vitro diagnostic medical devices and the provision of nucleic data analysis for human diagnostic.
On request we deliver sequencing analyses data with the requirements of GCP/GLP/GMP regulations.
Dr Andreas Ebertz
Did you like this article about NGS platforms and capacities? Then subscribe to our Newsletter and we will keep you informed about our next blog posts. Subscribe to the Eurofins Genomics Newsletter.














10 Comments