Exome sequencing is the preferred method to analyse single nucleotide polymorphisms (SNPs) and insertions and deletions (InDels). Especially clinical researchers are turning to exome sequencing to support and facilitate clinical diagnostics, treatment and prevention of disorders caused by genomic abnormalities.
However, some sample types can be quite challenging to sequence. Here, we focus on common clinical sample types and how to handle them for optimal sequencing results.
Sample types for human exome sequencing
The main sample types for exome sequencing are cells, tissue, formalin-fixed and paraffin-embedded (FFPE) tissue, and liquid biopsies.
For cells and tissue, a lot of established and reliable DNA extraction kits and protocols exist. In clinical settings, however, the starting materials commonly used for tumour mutation profiling are FFPE samples and liquid biopsies, from which cell-free DNA (cfDNA) is extracted.


What are the most decisive tips and advice that can help facilitate the efficient performance of exome sequencing of cfDNA and DNA extracted from FFPE samples?
cfDNA exome sequencing
A blood sample of a cancer patient, also called a liquid biopsy, contains cfDNA from non-cancerous cells and circulating tumour DNA (ctDNA). Significantly, the amount of ctDNA in a liquid biopsy tends to vary considerably between individual patients, the cancer type and the health status of the patient. It can be as low as 1% in the early stages of the disease. Moreover, DNA isolation from plasma results in very small amounts of total DNA, ranging from as little as 1 ng/ml to 10 ng/ml of plasma.
These factors show the critical necessity for highly sensitive processing and sequencing methods to achieve accurate variant calling.
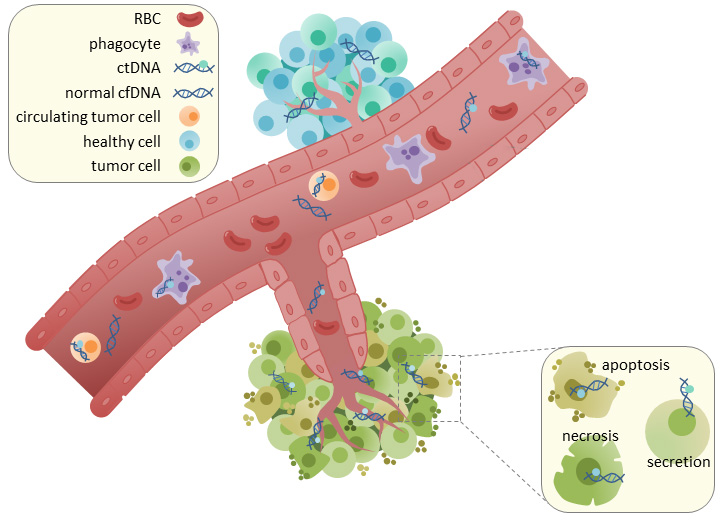
cfDNA sample preparation for exome sequencing
- Drawn blood should be collected in tubes that contain anticoagulant and preservative agents such as EDTA or citrate-phosphate-dextrose-adenine.
The blood samples should be stored at room temperature and the plasma needs to be prepared within 1 hour of being drawn. - Heparin should be avoided, as it may interfere with downstream processes.
- Blood stored in conservation tubes (Roche, Streck, Qiagen, etc.) should be stored according to the manufacturer’s instructions, which is usually a few days.
- Blood samples should never be frozen due to increased cell lysis and release of DNA.
- After preparation, plasma should be snap frozen in liquid nitrogen and stored at -80 °C. Shipment on dry ice if necessary.
In clinical investigations that are often quite large and extensive, the samples are the key element. Highly sensitive processing and sequencing will make or break the success of the investigation. Therefore, outsourcing these steps to excellently trained and experienced experts could be decisive. The Eurofins Genomics NGS experts have demonstrated again and again outstanding performances in both small and large projects.

For more details about best practise for cfDNA sample preparation, visit our sample preparation and shipping guide.
FFPE exome sequencing
The whole process of sample fixation and storage for FFPE samples, involving formalin and paraffin, can cause substantial DNA damage. Genomic DNA extracted from FFPE samples is often partially degraded or in very limited quantity.
The amount and type of damage in DNA extracted from FFPE material often varies extensively. Inter-and intra-strand crosslinks and an accumulation of strand breaks are common. FFPE material sometimes shows a low amount of C-to-T deamination artefacts, as well as other base substitutions.
These issues illustrate the importance of correct FFPE sample preparation for subsequent exome sequencing.
FFPE sample preparation for exome sequencing
- The tissue should be fixated as quickly as possible. Storage at 4°C should not be longer than a few hours as DNA starts to degrade. A quick fixation will also keep crosslinking and DNA fragmentation at a minimum.
- Before embedding, the samples should be completely dehydrated.
- Before DNA extraction, unstained and uncovered FFPE samples should be freshly cut into slices not thicker than 10 μm.
- After DNA extraction, the final DNA concentration should be higher than 2 ng/μl
- For exome sequencing, a total DNA amount should be at least 200 ng.
As the extraction and processing of DNA from FFPE samples can be very challenging, especially when it comes to the quality of the library, outsourcing these steps could be the best choice. The experts in our NGS labs process FFPE samples with established and validated protocols on a regular basis. The exome sequencing is performed with the sequencing depth to achieve accurate variant calling according to the project requirements.
For more details on FFPE sample preparation, visit our sample preparation and shipping guide, or get in contact with our NGS experts.
Library prep for exome sequencing
Besides appropriate sample preparation, the library preparation step is crucial for successful exome sequencing. In general, library preparations from FFPE and cfDNA samples are difficult. However, the utilisation of specifically optimised protocols for these sample types allows for a successful library prep.
The in-house library preparation steps at our NGS facility are performed with extreme caution in order to maximise the yield and quality of the genomic material. We only use high fidelity enzymes during this highly automated procedure.
Automation of the process enables consistent sequencing results and the execution of exome sequencing according to QM and QA standards.

Eurofins Genomics provides human exome sequencing with the highest possible coverage and unmatched prices. Find out more about our exome sequencing solutions.
Or get in contact with our NGS experts for more information about our exome sequencing capabilities.
Did you like this article about exome sequencing? Then subscribe to our Newsletter and we will keep you informed about our next blog posts. Subscribe to the Eurofins Genomics Newsletter.








