Key takeaways:
- NGS is used for disease biomarker discovery and biomarker validation.
- Whole exome sequencing has been used for diagnostics of poorly characterised diseases by identifying novel mutations and biomarkers.
- Whole genome sequencing is well-suited for diagnostics of rare diseases by identifying complex mutations such as chromosomal translocations.
- NGS is the pivotal system for the detection of diseases and the generation of personalised pharmacogenomics profiles.
NGS for biomarker discovery
The basis of diseases and disorders are found in genomic alterations. A stunning number of diseases and disorders, believed to number around 6,000 to 8,000 including sickle cell anaemia and lactose intolerance, are caused by variations in a single gene such as single-nucleotide polymorphisms (SNPs) (image 1). These diseases and disorders are inherited or caused by de novo events.
RNA sequencing, exome sequencing and whole genome sequencing are some of the preferred NGS choices to discover and validate biomarkers for diseases and disorders. For example, lncRNAs, miRNAs and mRNAs have been used as diagnostic biomarkers and even therapeutic targets for patients suffering from various diseases.
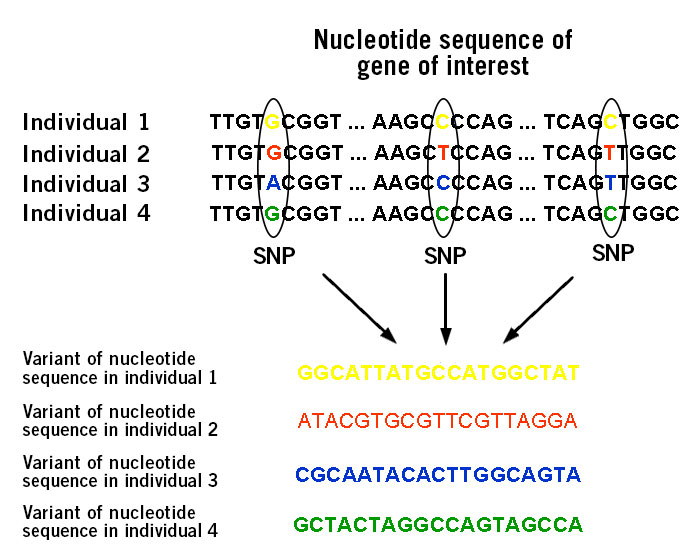
Image 1: Depiction of single nucleotide polymorphisms (SNPs).
NGS diagnostics: Range of diseases
For the diagnosis of complex diseases that involve multiple genes, NGS is the most suitable option. These diseases include autism, connective tissue disorders, cardiomyopathies (disease of the heart muscle that impedes blood pumping), and disorders of sex development (Di Resta et al., 2018). For instance, in 2009 Richard Lifton and colleagues at Yale University diagnosed the clinical condition of a patient using whole exome sequencing (WES) (exome = protein coding regions). The patient, a five months old infant, was suspected to have Bartter’s syndrome. However, the results of WES revealed the presence of a mutation that resulted in congenital chloride diarrhoea (Choi et al., 2009). The symptoms of both diseases are similar: intestinal problems and dehydration. In this and other cases, conventional genotyping methods such as karyotyping, fluorescent in-situ hybridisation (FISH) and restriction fragment length polymorphism analysis (RFLP) failed to properly diagnose the disease. The WES strategy opted for by Lifton was sheer brilliance and the decisive factor in finding the correct diagnosis, as around 85% of the disease-causing mutations are present in the exome. It also cut the costs for the diagnosis to a fraction of the other option of whole genome sequencing (WGS).
Image 2: Depiction of the personalised therapeutic approach.
Currently, “big NGS data” is utilised by some clinical laboratories for the development of personalised therapeutic approaches (image 2). This will help in understanding disease mechanisms, along with strategies to prevent and treat diseases. Soon clinicians will be able to utilise the patient’s genetic information to choose specific medical care that has the highest chance of success. Before this can be realised, legal, ethical and technological challenges need to be clarified by the respective authorities that, at present, are working on developing guidelines.
Nevertheless, there are three main NGS strategies for diagnostics purposes: NGS panels, WES and WGS. The choice of strategy depends on the nature of the individual disease and available funds.
NGS diagnostics by Exome Sequencing: For poorly characterised diseases
Only about 1% of the whole human genome encodes proteins. This part of the genome is called exome (image 3). Significantly, approximately 85% of disease-related mutations are present within the exon (the exome is composed of the entirety of exons). Utilising WES is an excellent approach when the genetic basis of a disease or phenotypic trait is rather poorly characterised. WES enables the identification of novel mutations and biomarkers for complex diseases such as psychiatric, autoimmune and neurodegenerative disorders. Primary immunodeficiencies, including chronic granulomatous disease, severe combined immunodeficiency and chronic mucocutaneous candidiasis, have already been diagnosed using WES (Arts et al., 2019).
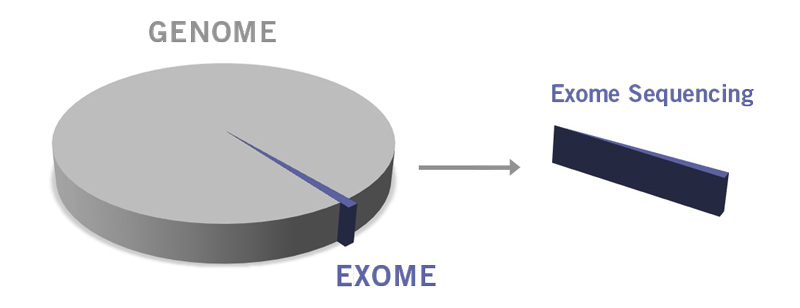
Image 3: Depiction of difference between genome and exome.
NGS diagnostics by Whole Genome Sequencing: For rare diseases and beyond
As the name indicates, WGS includes sequencing the coding (exons), non-coding (introns) and regulatory regions. WGS enables the identification of novel gene mutations including complex mutations such as chromosomal translocations and copy number variations (CNVs). It enables the diagnosis of rare disease that, when combined, affect more than 300 million people worldwide (Liu et al., 2019; Marshall et al., 2020).
As additional routine procedure in research laboratories, WGS is used to analyse infectious agents such as bacteria and viruses (Ilyas, 2017). Thus far, PCR-based genetic tests have usually been performed for viral infections but not for bacteria. Isolation, correct species identification and susceptibility testing of bacteria may take 1 to 2 days. But for slow growing strains, like Mycobacterium tuberculosis, phenotypic testing could take weeks. Additionally, less than 2% of bacteria are cultivable in laboratory conditions, which could complicate testing even more (Wade, 2002). In those cases, WGS is the preferred detection method. NGS also helps in analysing of the genetic changes that result in antibiotic resistances and, therefore, enable clinicians to prescribe effective antibiotics. For example, the genome of M. tuberculosis is homogenous and antibiotic resistances can arise through deletion, insertion or nucleotide substitution (Achtman, 2008; Ilyas, 2017).
Image 4: Rare disease affect around 300 million people worldwide and can be analysed by whole genome sequencing.
NGS clinical diagnostics: The challenges
NGS faces some challenges in clinical settings regarding the preparation of DNA/RNA samples, library preparation and read accuracy of sequences. In order to receive good sequencing results, the DNA sample should have a high yield, integrity and purity. This could be problematic when DNA samples come in formalin fixated and paraffin embedded form (FFPE) that have been stored for an extended period of time. Formaldehyde, the effective component of formalin, generates cross-linking between nucleic acids and proteins. During the fixation process, where a very low pH is present, fragmentation of the DNA takes place that, in turn, lowers the DNA yield and reduces its integrity (Gilbert et al., 2007). Sometimes, the amount of extracted DNA is less than the amount that is required for library preparation. The subsequent PCR amplification to increase the DNA amount inevitable introduces PCR bias. Optimisation of sample and library preparation procedures can mitigate these challenges.
PCR bias arises due to differences in the amplification efficiency of different templates or due to self-annealing of abundant templates and the resulting inhibition of amplification during the late amplification steps (Acinas et al., 2005).
After the sequencing run, the quick analysis of NGS data is paramount for diagnostic purposes. The process could be quite challenging and take a long time. However, various types of bioinformatics software have been developed and can be applied for sequence alignment to the reference genome, data analysis and subsequently correct diagnosis. (Gonzalez-Garay, 2014).
Overall, NGS as diagnostics tool can provide a more comprehensive view as well as many details of a disease that conventional detection methods are not able to gather.
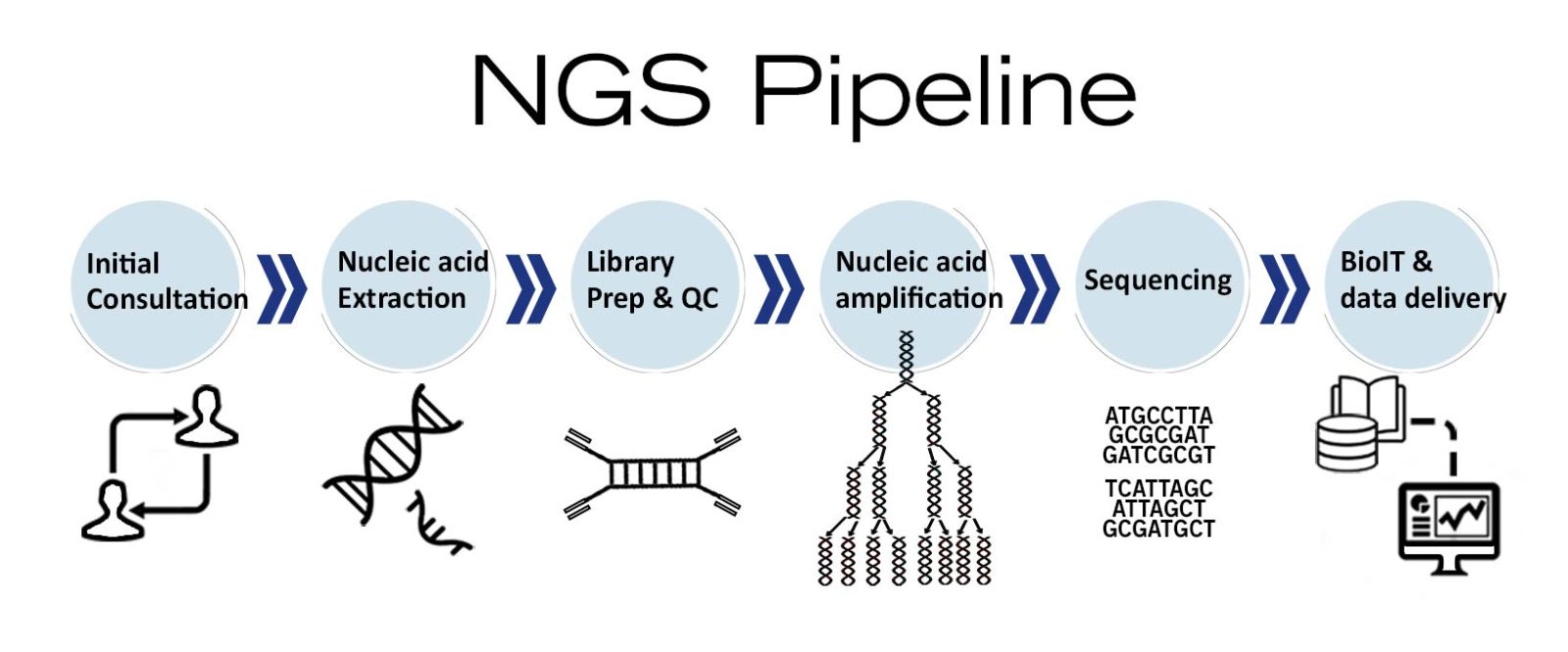
Image 5: Depiction of NGS workflow used as molecular diagnostics tool.
Personalised medicine: NGS takes medicine to the next level
Despite some challenges, NGS is paving the way towards personalised medicine! NGS has the potential to accelerate the detection of disorders and generate personalised pharmacogenomics (effect of genes on the individual’s response to drugs) profiles. After all, the genetic makeup of patients is not completely identical and different genetic mutations can cause the same disease and symptoms. Therefore, the correct diagnosis of a disease and taking into account the individual’s response to a therapeutic agent, which is genetically determined, is crucial. NGS can help in comprehending the biology of the disease and can assist in making clinical decisions based on an individual’s genetic profile. Ashley and colleagues at Stanford University School of Medicine, for example, sequenced the whole genome of a 40 years old healthy male patient with a family history of osteoarthritis, premature coronary artery disease and sudden death. The team focused on three types of variants: rare mutations, novel mutations and mutations that modulate drug response. Interestingly, the investigation found 63 known pharmacogenetic variants. The presence of these mutations suggested that this individual will respond well to statin while being resistant to clopidogrel. As both drugs are prescribed to prevent atherosclerotic cardiovascular disease, the findings of the study will help clinicians to plan individual prevention and treatment strategies for the patient if needed. Additionally, the investigation identified three rare mutations that are associated with sudden death, and another gene variant associated with coronary artery disease. These findings could also contribute to creating a personalised treatment plan for the individual in future (Ashley et al., 2010; Samani et al. 2010).
With the incorporation of NGS in clinical settings, the availability of genomic information will not be a limiting factor to guide and optimise therapies anymore. The dream of applying genetics to clinical medicine can finally become reality.
Are you interested in using NGS as a diagnostics tool or are you using NGS for your research? We make the use of NGS easy. Find out how on our NGS-made-easy page.
NGS is also successfully applied as clinical diagnostics tool in oncology.
The different types of NGS
NGS is offered in a wide range of applications that can be somewhat overwhelming. Find the NGS solution that fits your project best with our NGS decision tree.
By Tamseel Fatima and Dr Andreas Ebertz








2 Comments