Bacterial plasmids are peculiar genetic data carriers. They do not contain genes that are crucial to the bacteria’s survival per se. They rather contain genes that contribute to the bacteria`s thriving in specific environments, their pathogenicity and ability to utilise nutrients, but also contain antibiotic resistance.
Antibiotic resistance genes in plasmids are of particular interest in biology and medicine. In order to understand the steadily evolving mechanisms of antibiotic resistance, resistance genes have to be identified, analysed and their propagation in bacterial populations monitored.
Key takeaways:
- Plasmids can carry one or more antibiotic resistance genes. Together with transposons and integrons, plasmids are the main genetic elements responsible for the spreading of antibiotic resistance genes in a population.
- Whole plasmid sequencing provides high throughput sequencing data for the identification of novel antibiotic resistance genes and mechanisms.
- Whole plasmid sequencing is ideal for the characterisation of unknown plasmids and validation of known plasmids.
A well-known example for antibiotic resistance genes of bacteria are extended-spectrum beta-lactamase (ESBL) genes and beta-lactamases (BL) genes in E. coli. These genes are located on the plasmid and encode for enzymes that are able to inactivate beta-lactam antibiotics such as penicillin (image 1). Additional antibiotics resistances against tetracycline for instance could also be encoded on the plasmid. This complicates a treatment as E. coli containing ESBL / BL genes can survive a wide range of antibiotic exposure during medical treatments (Darphorn, 2021).
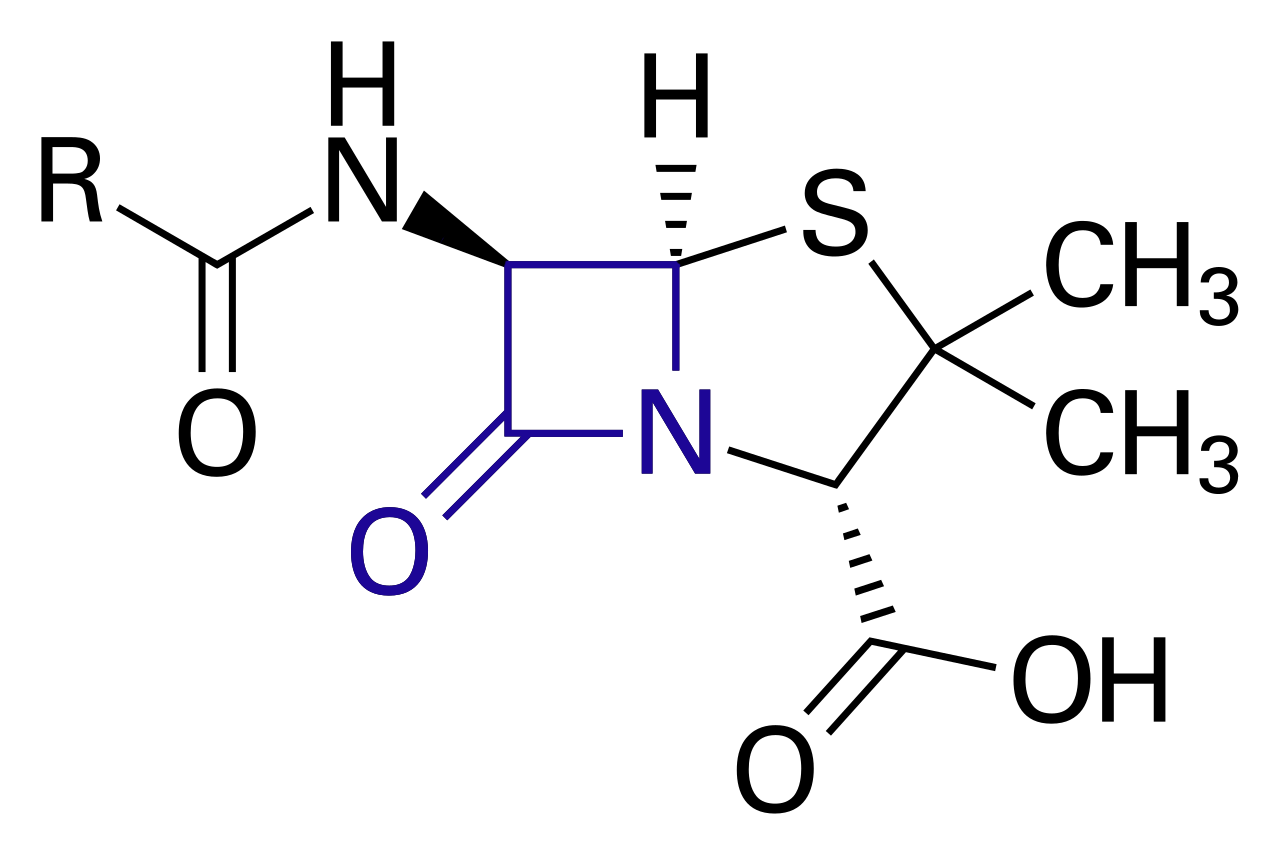
Image 1: Chemical structure of penicillin and target of beta-lactamase enzymes for penicillin degradation (in blue).
This and many other examples illustrate the importance of plasmids in antibiotic resistance and the need for complete sequencing of plasmids with high accuracy to gain more insights into the mechanisms of antibiotic resistance. Several methods for plasmid sequencing have been developed, however NGS-based whole plasmid sequencing often seems to be the most suitable method to characterise unknown plasmids and validate known plasmids.
Novel antibiotic resistance gene identification with whole plasmid sequencing
Whole plasmid sequencing facilitates the characterisation of plasmids, as it provides sequencing data of the complete plasmid. This data is then used in comparative analyses with known plasmids to identify gene variations in resistance genes. Here, the identification of clusters with multiple resistance genes, called resistance islands, are especially interesting for the discovery of novel resistance genes and other determinants involved.
A functional annotation analysis of the gene variations could then predict and even indicate genes involved in novel resistance mechanisms. In a following functional validation, the potential resistance genes are cloned into host bacteria and their impact on the susceptibility of the bacteria to antibiotics is assessed.
Analyses of the sequence and structure of the potential resistance genes combined with the experimental data could elucidate the mechanisms involved, eventually paving the way to the development of novel antibiotics and therapeutic strategies.
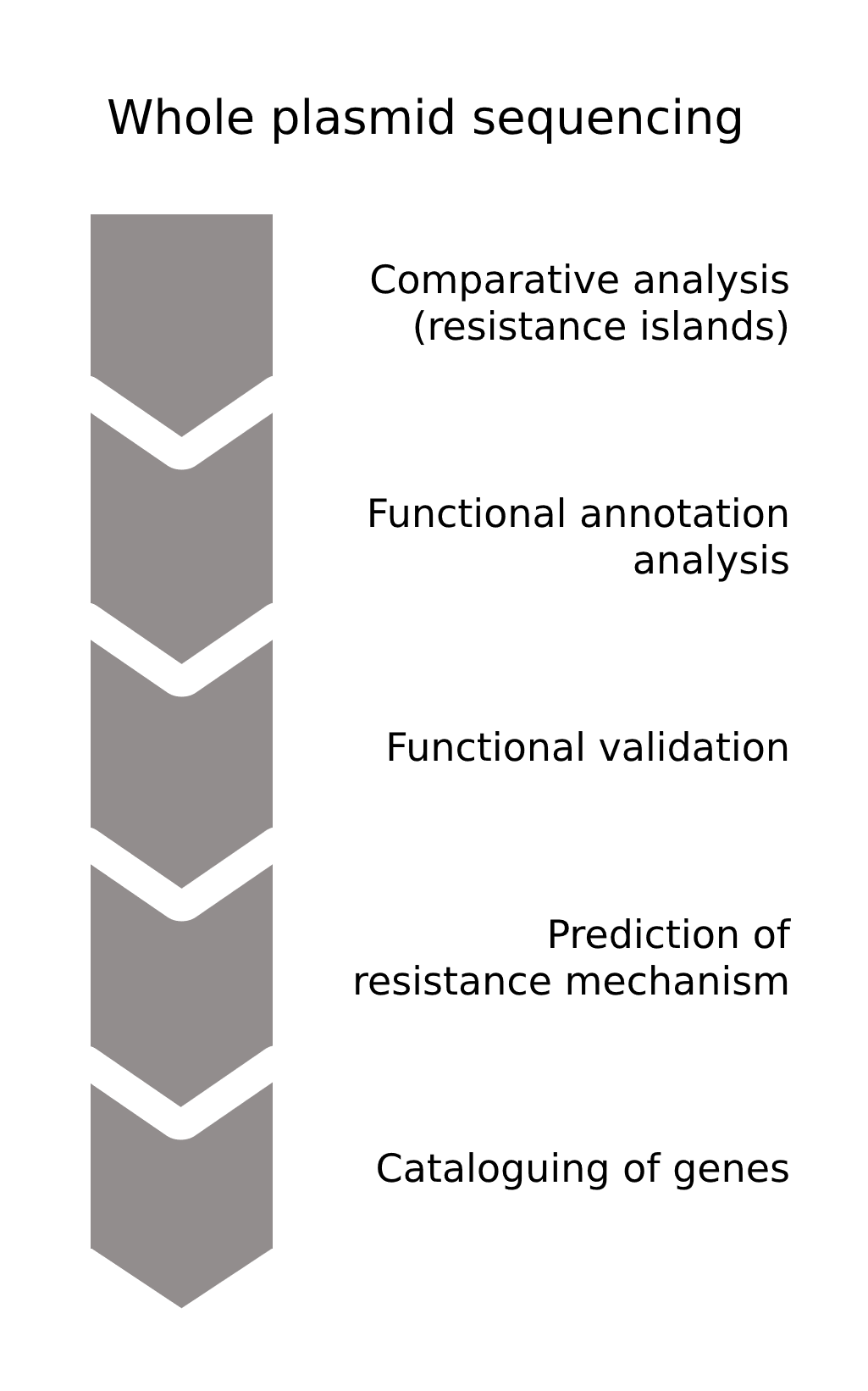
Image 2: Depiction of the process of identifying, validating, and analysing novel antibiotic resistance genes and mechanisms (process simplified).
The basis for the in-silico analyses are comprehensive databases on bacteria that contain all genes, specifically known and potential antibiotic resistance genes; databases such as The Comprehensive Antibiotic Resistance Database (CARD), ResFinder and AMRFinderPlus. NGS-based whole plasmid sequencing increasingly contributes to the pool of data (Feldgarden et al., 2021; Schmartz et al., 2022).

Image 3: Schematic representation of a plasmid with backbone, resistance genes, Promoter and origin of replication.
Whole plasmid sequencing and transposons / integrons
Transposons are mobile genetic elements that can “jump” to different locations within a genome but also plasmids in bacteria. Transposons can transfer between plasmids or from chromosome to plasmid and vice versa. It has been found that transposons play a role in antibiotic resistance as they can contain antibiotic resistance genes against beta-lactams, aminoglycosides, and others. Some transposons have been found to induce conjugation and their own transfer into another bacterium (conjugative transposons). Additionally, plasmids could encode enzymes involved in transposition of transposons (transposase).
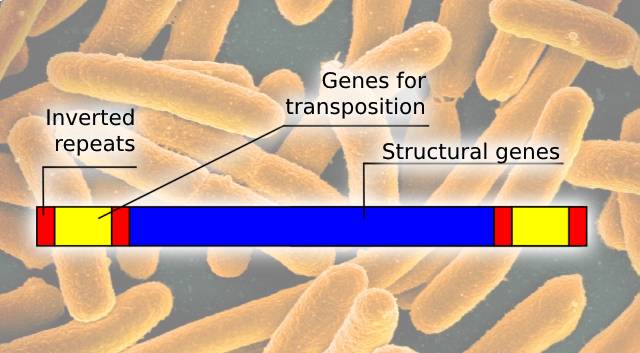
Image 4: Schematic representation of a bacterial transposon with structural genes, genes for transposition that could include genes encoding for antibiotic resistance, and inverted repeats. Bacterial transposons are DNA transposons and belong to the Tn family (1).
Integrons are also genetic elements and can carry different gene cassettes “accumulating” multiple antibiotic resistance genes over time. Integrons such as class 1 integrons that are present in a high amount of gram-negative pathogens can be transferred to other bacterial pathogens, therefore, contribute to the formation of multidrug-resistant bacterial populations.
Overall, plasmids, integrons and transposons are the main elements of spreading antibiotic resistance genes via horizontal gene transfer. Whole plasmid sequencing provides data for the complete plasmid, thus, facilitating the analysis and understanding of these genetic elements. After all, it has been proposed that blocking the transposition of these genetic elements could open new ways to reduce the spread of antibiotic resistance (Gillings et al., 2008; Pérez-Valdespino et al. 2016; Babakhani and Oloomi, 2018; Rubio-Cosials et al., 2018).
Whole plasmid sequencing for plasmid tracking
Whole plasmid sequencing is also used for tracking and monitoring of specific bacterial species in epidemiological investigations. Here, plasmid sequences are utilised as molecular markers for tracking the transmission of specific bacteria. In fact, the transfer and evolution of antibiotic resistance genes on plasmids have been monitored using whole plasmid sequencing in a recent study. The technology enabled the researchers to investigate the emergence of multidrug-resistant bacteria (mediated by the transfer of a multidrug resistance plasmid) in clinical and environmental samples. Furthermore, they proposed the utilisation of whole plasmid sequencing for infection control in real-time, leading to a more efficient containment of outbreaks in clinical settings (Peter et al., 2022).
At Eurofins Genomics, we use Nanopore technology for whole plasmid sequencing.
Nanopore technology is based on the detection of changes in electrical conductivity during the passage of DNA strands through pores the size of nanometres.
Get whole plasmid sequencing starting from 15 € per sample (other currencies applies).