A programmable yeast cell factory for tuneable fatty acid-based production
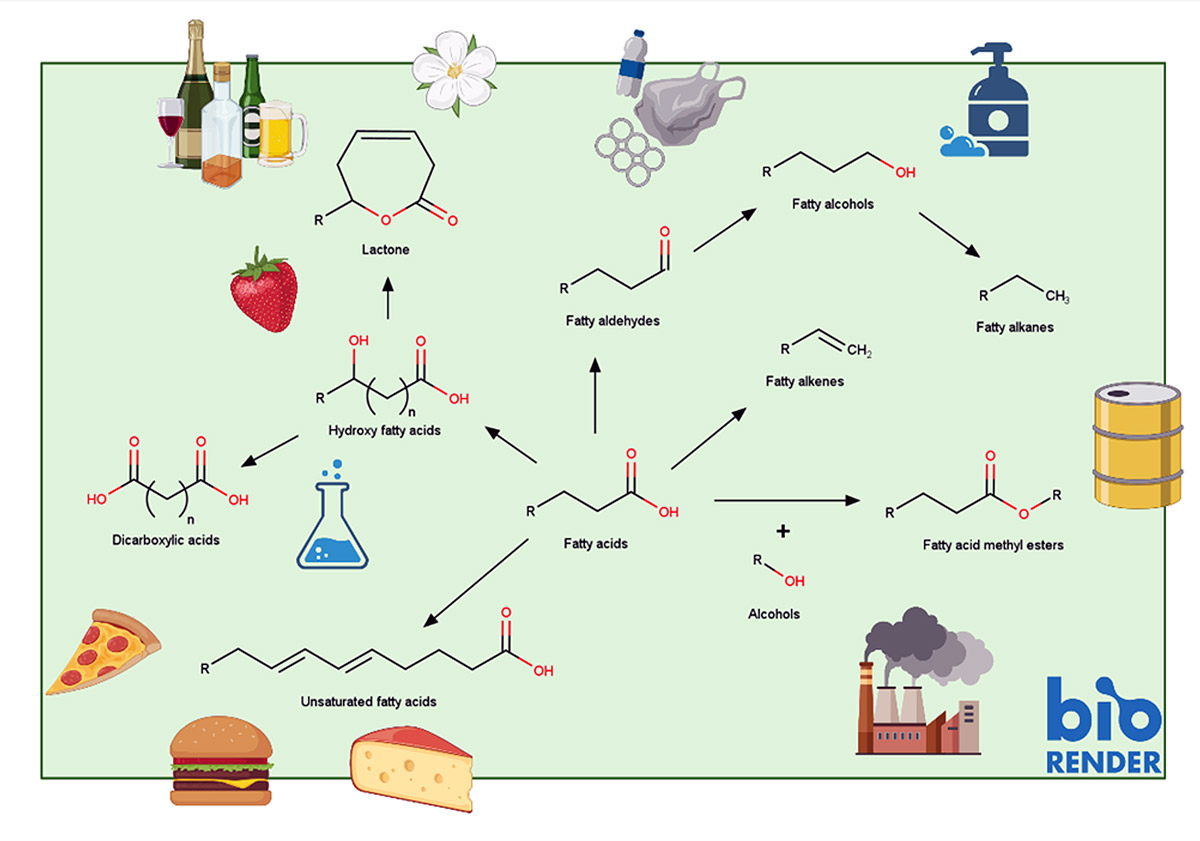
Fatty acids are essential for biological life, and they are also pivotal to modern life. Fatty-acid derived compounds called oleochemicals such as hydroxy fatty acids, fatty alcohols, fatty acid methyl/ethyl esters, and fatty alka(e)nes are key building blocks for many industrial products. Their physiochemical properties allow for a wide range of chemical reactions to be performed, and applications can be found in diverse areas such as fuels, pharmaceuticals, detergents, lubricants, coatings, plastics and nutraceuticals.
Image 1: Showing examples of oleochemicals ubiquitous use in the production of industry- and commercial products. Specific application areas and physiochemical properties are heavily dependent on both the type, and the chain-length of the foundational fatty acids. For a more comprehensive overview of fatty acid chemistry please have a look at the full project description which will be published on the iGEM Wiki page from Team:Chalmers-Gothenburg at the beginning of November 2021. Modified from Yu et al., 2014 (open-access article distributed under the terms of the Creative Commons Attribution License).
The challenge of sustainable fatty acid production
Environmental protection and recovery are major topics in the 21st century and there is now an urgent need to find alternative manufacturing methods for oleochemicals, as this would significantly boost our efforts towards a sustainable bio-based economy. The global fatty acids market is forecast to reach 48.46 billion USD by 2027 (GlobeNewswire, 2020). While plant oils have largely replaced crude oil as the raw material for many oleochemicals, the production methods of plant oils such as palm oil are major drivers of deforestation and biodiversity loss through excessive land and water use, as well as long-distance transportations. Furthermore, conventional manufacturing processes tend to be energy inefficient (requiring high temperatures and pressures) and involve large quantities of acids, bases, and organic solvents.
Yeast cell factory: The future of microbial factories
An alternative source of oleochemicals is engineered microbes. Much effort has already been invested on metabolic engineering of high-yield microbial strains; however, these are often limited to a smaller number of structurally simple, low-molecular weight products such as aromatics, amines, terpenoids, terpenes and esters (Calero & Nikel, 2019; Navarrete et al., 2020). Synthetic biology is now bringing new tools and approaches to developing microbes as programmable platforms that allow the production of a broader range of compounds. By designing and introducing completely novel metabolic pathways, scientists can produce compounds that previously could only be effectively isolated from natural sources such as complex opioids and vitamins (Galanie et al., 2015; Sun et al., 2019).
Significantly, microbial-based manufacturing processes also have the potential to address many of the challenges associated with plant-based supply chains. Industrial cultivation of many microorganisms occurs over days, whereas plant production is often annually and regionally limited. As microbes can be grown in closed systems such as bioreactors, the production process is not challenged by external environmental factors. In comparison with oleochemicals based on plant oils, cell factories potentially allow greater consistency in product composition and purity profiles. Plant-based systems produce a mix of different acyl chain lengths, which affects the properties of the final product (Carlsson, 2009).
Utilising a yeast cell factory could result in a stable, controlled, and tuneable production of oleochemicals. After all, the fatty acid chain lengths are important determinants for the properties of the final product.
Using a yeast cell factory to set the fatty acid chain lengths
The model organism Saccharomyces cerevisiae has been proven to be a reliable platform for industrial large-scale production of various compounds due to its robustness, genetic malleability, and tolerance towards harsh fermentation conditions (Parapouli et al., 2020). In the yeast fatty acid synthesis, the substrates enter the fatty acid synthase (FAS) complex, a large barrel-shaped protein complex (2.6 MDa) (image 2). The yeast FAS complex has proven to be a challenging target for engineering due to its cooperative nature, as the function of the complex is encoded in large protein blocks, rather than individual enzymes. Thus, a replacement of the yeast FAS complex with a system more accessible to engineering efforts may appear evident. The bacterial type II FAS consists of discrete monofunctional enzymes that can be manipulated individually, including the thioesterase responsible for the final chain-length (image 3) (White et al., 2005). Fernandez‐Moya and colleagues (2015) demonstrated that the functional replacement of the S. cerevisiae FAS with the Escherichia coli FAS (eFAS), together with the integration of heterologous thioesterases, allowed for flexible production profiles of fatty acids with different carbon chain lengths (Fernandez‐Moya et al., 2015).
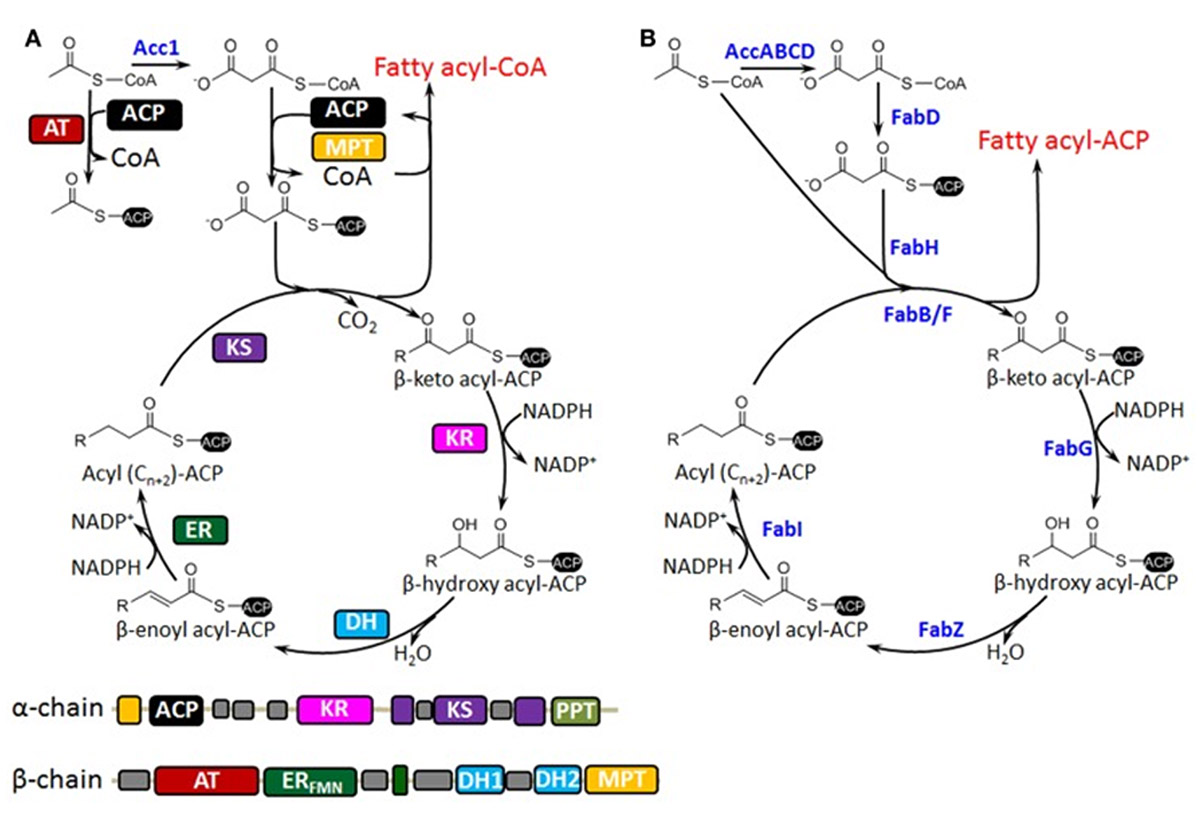
Image 2: Showing a comparison between S. cerevisiae type I (FAS) (A) and E. coli type II (eFAS) (B) fatty acid synthesis. In FAS, the catalytic domains are located on the interconnected α-, and β-chains. In eFAS, the similarly performing enzymes are distinct and monofunctional (marked in blue). In both systems, fatty acid synthesis in initiated by acetyl-CoA, the starting point for a new chain capped by a CoA cofactor. Malonyl-CoA is then iteratively fed into the reaction cycle to continue building the chain by two extra carbon atoms at a time. The process stops when the terminal CoA unit is cleaved off by a thioesterase. Most thioesterases target certain ranges of chain lengths. The overall fatty acid profile that is produced by the cell is then dependent on which thioesterases are expressed. Image and text modified from Zhou et al. (2014) (distributed under the terms of the Creative Commons Attribution License).
Our project: Designing a yeast cell factory
In our project, we investigate the possibility of producing different oleochemicals with specific compositions by regulating the chain lengths of the fatty acids used as starting compounds. We want to explore if it is possible to regulate the expression levels of the thioesterases through different induction systems, and in combination with various enzymes, catalyse the fatty acids into high value oleochemicals such as fatty alcohols, esters, and alkanes. By fine-tuning the fatty acid profiles, we aim to influence the physiochemical properties of these compounds, as they are heavily dependent on the alkyl chain length.
To test our hypothesis, we integrate the eight eFAS genes from E. coli K-12 MG1655 (acpP, acpS, fabB, fabD, fabG, fabH, fabI, and fabZ) into the genome of S. cerevisiae CEN.PK 113-11C. We then link three thioesterases with different chain-length preferences (TesA from E. coli, FatB from Ricinus communis, and TesBT from Bacteroides thetaiotaomicro) to three chemical induction systems. Inducers are molecules that regulates gene expression and they function by either binding to activator proteins that help to recruit RNA polymerase or disabling repressors that prevent transcription. To provide a proof-of-concept we chose to work with (1) the CUP1 system which is native to yeast (where it responds to copper by producing metallothioneins), (2) a tetracycline system based on bacterial antibiotic resistance genes, and (3) an estradiol system based on the mammalian hormone β-estradiol (Bellí et al., 1998; Labbé & Thiele, 1999; McIsaac et al., 2011). To find optimal concentration ranges, we benchmark each system individually, in pairs, and as combination of all three systems, by replacing the thioesterase genes in each system with different fluorescent proteins (GFP, RFP and BFP).
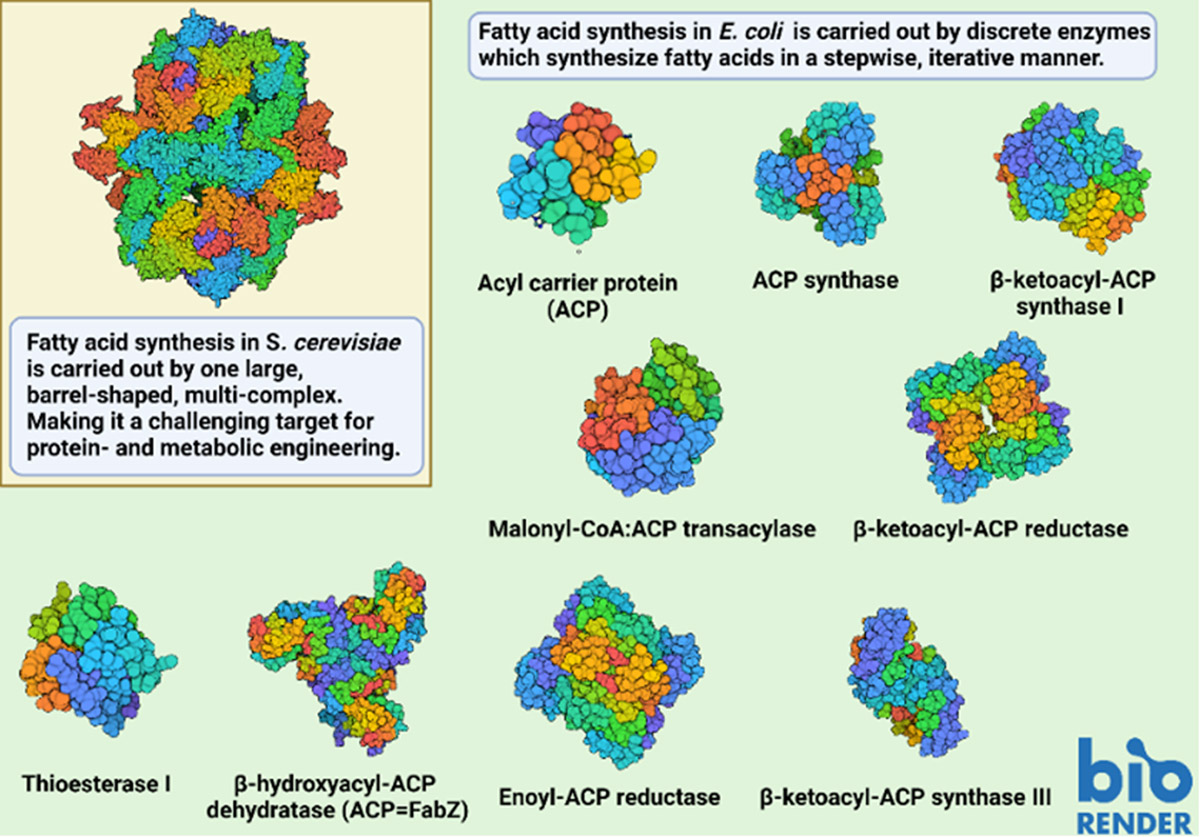
Image 3: Showing the structure of the FAS complex in S. cerevisiae (PDB 6TA1) in comparison with the nine enzymes involved in E. coli eFAS (PDB 1T8K; 5VBX; 1FJ4; 2G2Y; 1I01; 1HNK; 5CFZ; 6N3P; 5TIC). Visualising complexes of ACP with its enzymatic partners has been a challenging research effort as many complexes are short-lived. Dodge and colleagues (2019) overcame this problem by employing cross-linking probes in combination with protein fusion methods to covalently trap E. coli FabZ in functional association with ACP to solve the crystal structure of the resulting complex (ACP=FabZ) (Dodge et al., 2019).
Yeast cell factory: The use of multiple signals allows programmability and fine-tuning of production
Our team propose that S. cerevisiae can be engineered to allow the fine-tuning of reaction pathways in vivo by localisation and compartmentalisation of the enzymatic reactions that perform each step of the reaction and utilising more sophisticated induction- and signalling systems. Establishing synthetic pathways for the production of biochemicals is often hampered by crosstalk between competing pathways, the production of undesirable and/or toxic side-products and cellular responses, while co-localising enzymes by using synthetic scaffolds has been proven to improve production. Here, the concentration and compartmentalisation of intermediates in cellular organelles such as vesicles can isolate the pathways from any competition and provide a more suitable environment for biosynthesis (Dueber et al., 2009; Lee et al., 2012; Zhou et al., 2016; Reifenrath et al., 2020; Dickey et al., 2021). We would like to highlight the excellent work done in this area by the iGEM 2017 Team:Cologne-Duesseldorf, who designed tools for customising cell compartments in yeast by regulating the import of enzymes into peroxisomes.
Precise and accurate regulation of gene expression may be considered as the ‘Holy grail’ of synthetic biology. Exciting new technologies utilising synthetic GPCR signalling transduction pathways and dCAS-protein fusions with various gene regulatory domains enable the fine tuning of expression in a very precise manner (Doudna & Charpentier, 2014; La Russa & Qi, 2015; Billerbeck et al., 2018; Lengger & Jensen, 2020; Shaw et al., 2019). Facilitation of both ON/OFF states and the dynamic regulation needed to generate concentration ranges promise a route to generate a vast library of oleochemicals, even those difficult to extract or not found in nature. The use of a system like this could contribute to eliminating the need for environmentally harmful catalysts and solvents and could serve as a platform for new exciting (bio)chemistry!
Team:Chalmers-Gothenburg
Our team consisting of eight undergrad students from Chalmers University of Technology and University of Gothenburg. We are mainly supported by the Siewers Lab at the Division of Systems and Synthetic Biology (Chalmers). Our project is conducted as part of the 2021 International Genetically Engineered Machine (iGEM) competition. The iGEM Foundation is dedicated to the advancement of synthetic biology through the development of an open and collaborative community. Through an annual international competition iGEM support student researchers to conduct their own synthetic biology projects.

Group Picture: Presenting the members iGEM 2021 Team:Chalmers-Gothenburg. Clockwise from the top we have Fanny Linnéa Carlsson, Lina Andréasson, Erik Gingsjö, Olof Dahlman, Robin Nilsson, Xi Chen, Christer Edvardsson, and Edwin Eliasson.
Please do not hesitate to reach out to us with any questions regarding our project through our email (igemgothenburg@gmail.com). A more detailed project description will also be available at our Wiki page which will be complete at the end of 2021. We would also like to recommend that you visit our blog – The Transcriptome. The blog is co-written with Team:UNILausanne with the goal of making synthetic biology more accessible to those working outside the field.
Find out about other iGEM projects of 2021:

How can the microbiome be transformed to cure or treat diseases? The iGEM Team Heidelberg tries a different approach.

How can chloroplasts be used to establish a process to produce cell-free systems? Read how the iGEM Marburg team aims to shape a green future!

The iGEM TU Kaiserslautern is establishing modular cloning in Leishmania tarentolae for human-like-protein production.
By Christer Edvardsson and Dr Andreas Ebertz
Did you like this article on programmable yeast cell factory? Then subscribe to our Newsletter and we will keep you informed about our next blog posts. Subscribe to the Eurofins Genomics Newsletter.