OpenPlast – The chloroplast holds the key to agriculture and modern crop development
Since the dawn of mankind, agriculture has been essential for the development of human civilisation, and the genetic modification of crops to suit our needs is just as old.
Take corn, for example. Corn cobs used to be a lot smaller and held much fewer grains than the commercial corn we cultivate today. This improvement in crop yield has been achieved through millennia of selective breeding. Nowadays, however, we have powerful tools at our disposal that enable the targeted modification of the plant’s DNA. Such tools make it possible to give plants whole new characteristics, going beyond the slow pace of selective breeding. The power to reshape organisms at will demands great responsibility. This year’s iGEM Project from the Marburg University aims to facilitate the development of better, more responsible crops.
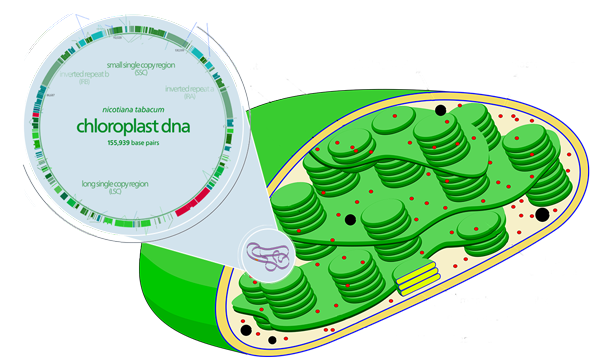
Most genetically modified crops nowadays rely on editing the DNA inside the cell’s nucleus; however, the nucleus is not the only place where DNA is present in the cell. The chloroplast, for example, is a very interesting engineering platform with several advantages over the plant nucleus.
The role of chloroplast in plant engineering
Chloroplasts can be engineered with very high precision, thanks to homologous recombination. In addition, transgenes can be stacked in operons – a trait that comes from the chloroplasts bacterial origin. Moreover, the chloroplast DNA lacks epigenetic silencing of transgenes, a mechanism present in the plant nucleus that can silence genes that have been inserted.
Last but not least, and probably the most noteworthy trait of chloroplasts: they are inherited maternally, which means that one cannot find chloroplast DNA in pollen. This drastically reduces the risk of unwanted transgene transmission if chloroplast-engineered plants were to be planted on our fields. This is an important aspect to the biocontainment of GMOs, a topic which is becoming more and more relevant – especially for the public acceptance of GM crops.
But if chloroplast engineering is so great, why aren’t there any crops with transgenic chloroplasts (transplastomic plants) currently on the market?
To answer this question, we contacted researchers working for key players in the industry, and their response was that current methods for creating transplastomic plants are too laborious and time consuming, making the effort not worth the risk: chloroplasts per se might be easier to understand and engineer, but getting the recombinant DNA into the chloroplast remains a challenge. Furthermore, chloroplasts contain multiple copies of their genome and making sure all of them have been edited requires months of selective breeding – a huge hurdle for scientists.
The OpenPlast project: Speed up the testing of chloroplast synthetic biology
This is where the OpenPlast Project comes in. We are working on a cell-free approach to speed up the testing cycle of genetic parts in chloroplasts.
Cell-free technology works on the principle that one can isolate cells (or organelles), break them down, and use their insides to test genetic constructs as if they were alive. This removes the need to have to bring DNA inside the cell – just pipette the DNA into the cell-free system (CFS) and watch while the cell’s transcription and translation machinery produces the desired protein in vitro.
The main drawback of characterising genetic parts in chloroplasts is the long months it takes for the plants to grow. OpenPlast’s chloroplast cell-free system can be used as a prototyping tool to test new genetic elements at an unprecedented speed and scale!
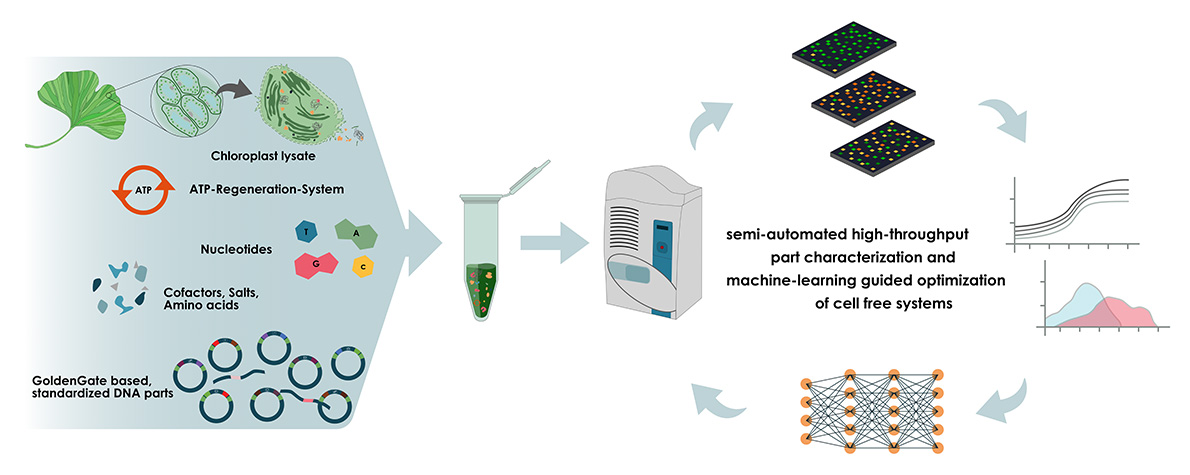
Instead of plant-filled greenhouses and months of painstaking work, we want to create a future where biologists are able to test hundreds of prototypes in a matter of hours (image 3)!
Image 3: Graphical abstract of the OpenPlast project.
In order to provide efficiently working extracts, we are using a machine-learning guided approach to optimise the composition of our cell-free systems. Concentrations of all components are varied in thousands of reactions, identifying the key components that influence expression in our systems the most. This should not only provide us with reliable working extracts, but generate valuable data that can be used by other scientists to optimise their own cell-free systems based on our findings!
A lack of tools & The Marburg Collection V.3
Nonetheless, even if we develop such an optimised system, we still have to show that they can actively be utilised for reliable characterisation of genetic parts. Genetic parts are small DNA elements that have a certain function, e.g. to start or stop transcription of a certain gene, and can be combined to create larger genetic sequences.
The amount of such genetic parts that can be used to engineer the chloroplast is very limited. In the iGEM registry, one of the largest part-collections world wide, contains more than 20,000 genetic parts, but only 18 of them have been designed to be used in plant chloroplasts.
This problem is well-known beyond the iGEM community. For rice, there is currently just one chloroplast promoter available (Lee et al., 2012), which shows the urgent need for more well-characterised parts in this field.
In 2018, the iGEM Team Marburg started a genetic part collection termed “The Marburg Collection”, a Golden Gate DNA Assembly based cloning toolkit for Vibrio natriegens. Finally, this collection is soon to be published (Stukenberg et al. 2021) and was expanded in 2019 with more parts for the use in cyanobacteria. Check out their article on our blog for more details on how this cloning system works!
The parts we are currently constructing for our project will comply with the framework of the Marburg Collection and expand its use to the chloroplast of various plants, hopefully fuelling further research in this exciting field.
In vivo vs. in vitro
To further demonstrate the use of our cell-free systems as prototyping platforms, we want to ensure that measurements in our CFS are actually comparable to in vivo data.
As chloroplast transformations are a tedious and time consuming process, we will be focusing on two major model organisms, Nicotiana tabacum and Chlamydomonas reinhardtii.
We aim to show that genetic regulatory elements characterised in our CFS will still perform similarly in these organisms. We are aware that we cannot count on absolute values to be the same, but in relation to each other, they should still show the same tendencies, e.g. a promoter shown to be stronger than others in CFS should still be stronger in the actual organism.
Outlook
Engineered plants will play an important role in our future. The changes could lead to improved yield or nutritional value, more resilient crops in the face of the drastic climate changes, or even for more sophisticated projects, like improved CO2 fixation.
We are OpenPlast and we want to help shape this green future.
Want to know more? The iGEM Team Marburg is always happy to answer questions via email (igem2021@synmikro.uni-marburg.de) or contact them on Twitter and Instagram @igemmarburg.
By Jonas Freudigmann and Yasoo Morimoto
Did you like this article on chloroplast DNA? Then subscribe to our Newsletter and we will keep you informed about our next blog posts. Subscribe to the Eurofins Genomics Newsletter.