Population genetics studies how specific genetic traits are passed on from generation to generation. They are applied in fields such as breeding research. Here, population genetics studies aim to identify genes that could be utilised as genetic markers for selection and calculate effective population sizes, but also to understand the evolution of pathogens and pests in order to establish control measures (Waples et al., 2016).
In humans, population genetics is often used to investigate the genetic basis of disease susceptibility, and further progress into the direction of personalised medicine.
Key takeaways:
- Population genetics studies in humans aim to identify genetic markers for a range of applications such as disease susceptibility.
- Genetic variations have been associated with increased risk for breast cancer, Alzheimer’s disease, type 2 diabetes, and high cholesterol levels.
- Population genetics studies contribute to the Polygenic Risk Score (PRS) – an estimation for disease susceptibility based on an individual’s genetic profile.
- Population genetics studies are used in pharmacogenetics to determine the effects of genetic variations on drug metabolism and adverse drug reactions.
Population genetics drive disease research
A well-known example for the effect of genetic variation of disease susceptibility are BRCA1 and BRCA2 for breast and ovarian cancer. BRCA1 and BRCA2 encode for proteins involved in DNA repair and are also called tumour suppressor genes. Large studies with clinical information from thousands of patients have found that people with certain genetic variations and mutations in BRCA1 and BRCA2 have increased risks of several cancer types – mainly breast and ovarian cancers but also pancreatic cancers (National Cancer Institute, 2020; Momozawa et al., 2022).
In Alzheimer’s disease research, it has been found that Apolipoprotein E (APOE), a gene involved in cholesterol and fat transport in the blood, is associated with Alzheimer’s disease. Similar to BRCA1 and BRCA2, individuals have two copies of APOE, one copy inherited from each parent. The presence of one or two copies of the gene variant APOE ε4 is linked to increased risk of Alzheimer’s disease. In fact, the presence of APOE ε4 is the strongest genetic risk factor for accelerated cognitive decline and late onset Alzheimer’s disease (Abondio et al., 2019; Gharbi-Meliani et al., 2021).
Population genetics dives deeper into genetic variation types
Disease risk can be determined by a range of genetic variations, some more common and some rare variations. Common genetic variations represent the larger proportion of the population of variations, whereas rare variations occur less frequently. Nonetheless, rare variations could have a stronger impact on disease risk.
It has been found that the risk for type 2 diabetes is increased in individuals with a specific variation in the TCF7L2 gene (encodes for a transcription factor). This specific genetic variation is rather common in populations of variants and is the most potent genetic risk factor for type 2 diabetes. On the other hand, a rather rare variation of the PCSK9 gene (Proprotein Convertase Subtilisin/Kexin type 9) has been linked to exceptionally high cholesterol levels and atherosclerosis (Grant, 2019; Guo et al., 2020; Del Bosque-Plata et al., 2021).

Population genetics for polygenic risk scores
Population genetics studies have been used to establish the polygenic risk score (PRS), an estimation of an individual’s susceptibility to a disease or predisposition to a trait based on their genes and variations therein. Here, large-scale genome-wide association studies have found gene variations associated with diseases such as coronary artery disease. In fact, more than 150 loci associated with coronary artery disease have been identified (coronary artery disease is the most common type of heart disease). It has also been found that the PRS for coronary artery disease could predict recurrent cardiovascular events in individuals with pre-existing coronary artery disease. Even though, PRS for coronary artery disease is still mainly in the research & development stage, the results are promising that it is suitable for clinical application. The utilisation of PRS could facilitate early intervention and the development of personalised prevention strategies (Levin and Rader, 2020; O’Sullivan et al., 2022; Fahed and Natarajan, 2023).
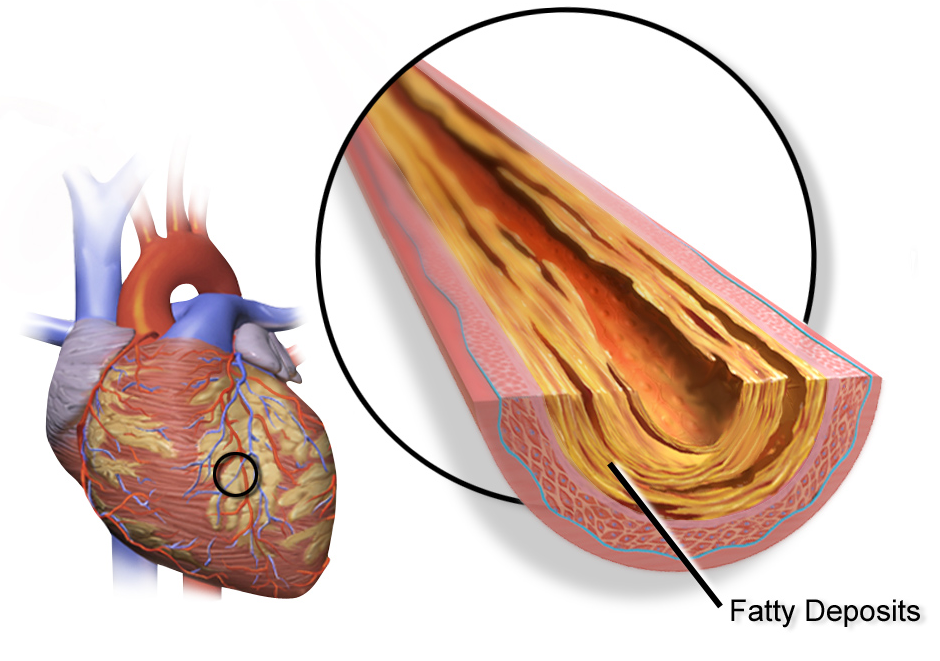
Image: Visualisation of coronary artery disease, where it can occur (left) and fatty acid deposition resulting in constriction (right) (1).
PRS could also substantially contribute for breast cancer risk prediction, screening, and preventive strategies. It has been estimated that around 30% of familial heritability of breast cancer, the most common cancer in women globally, can be attributed to gene variations, specifically certain single nucleotide polymorphisms (SNPs). The integration of PRS into risk models could facilitate the identification of women at higher risk of breast cancer development. In the future, PRS could also provide information important for the choice of screening frequency for women with increased risk and improve treatment strategies. Early detection of breast cancer in particular is of utmost importance for survival (Ginsburg et al., 2020; Ayoub et al., 2023; Roberts et al., 2023).
A fast and low-cost approach to identify genetic variations such as specific SNPs for the calculation of PRS could be the utilisation of SNP microarrays.
Population genetics drives pharmacogenomics
Pharmacogenetics studies the effect of genes on an individual’s response to a drug, and, thereby, guiding decisions on drug and dose selection.
Population genetics studies play a pivotal part in pharmacogenetics. In these studies, genetic variations are found that affect drug metabolism, efficacy and occurrence of adverse drug reactions. A widely known example for genetic variations are SNPs in the gene encoding for the enzyme Cytochrome P450 2D6, CYP2D6. These genetic variations result in varying enzyme activity and metabolization of a broad range of drugs (Monte et al., 2018).
Another important field of study are adverse drug reactions. It has been estimated that adverse drug reactions occur in over 30% of drug prescriptions! Genome-wide association studies have found more than 400 genes that are linked to drug efficacy and safety, as well as more than 200 genes linked to adverse drug reactions. Broadening our understanding of the interaction of gene variations and adverse drug reactions could minimise and mitigate adverse effects and complications (Cacabelo et al., 2019).
Population genetics studies greatly contribute to the identification of genetic markers for increased susceptibility to diseases such as breast cancer, Alzheimer’s disease, type 2 diabetes, and atherosclerosis. With advancing technology, population genetics will increasingly impact the new fields of precision medicine and personalised medicine.
Find out more about genetic biomarker validation by next generation sequencing (NGS), microarrays, qPCR/dPCR and Sanger sequencing.
Find out about the use of NGS as tool for clinical diagnostics of cancer, autoimmune and neurodegenerative disorders.
Did you like this article about population genetics? Then subscribe to our Newsletter and we will keep you informed about our next blog posts. Subscribe to the Eurofins Genomics Newsletter.