Microbiome transformation: How to train your gut microbiome for health
It has been established that the gut microbiome plays an essential role in our health. Several diseases, especially diseases that affect the gastrointestinal tract such as inflammatory bowel disease, have been associated with a dysbiosis of the microbiome.
Microbiome transformation: Prior therapies for the gut microbiome and their problems
An already established treatment method in this field is the so-called faecal microbiota transplantation (FMT), better known as stool transplantation. It involves transferring the stool of a healthy donor into the intestine of a diseased person through endoscopy or in the form of capsules. However, since the onset of the Covid-19 pandemic, FMT has been almost completely suspended. The risk of infection with Covid-19 through faecal-oral transmission cannot be ruled out.
Our solution for microbiome transformation: Personalised treatment of your own gut microbiome
We, the iGEM Team Heidelberg, are working to contribute to finding a solution to this problem. Our goal is to develop a method to treat microbiome-related diseases. In the future, our method should be capable of treating diseases without changing the composition of the microbiome.
For this purpose, we make use of specific bacteria that possess natural competence. These bacteria can take up plasmids naturally, without using established transformation protocols in lab settings such as “heat shock” or “electroporation” (Biggs et al., 2017).
We aim to develop a capsule that contains plasmids with the gene of interest and transports them to the intestine. Intestinal bacteria will then take up these plasmids by natural transformation and express the gene of interest (figure 1). The product of this gene is expected to specifically cure diseases or alleviate symptoms.
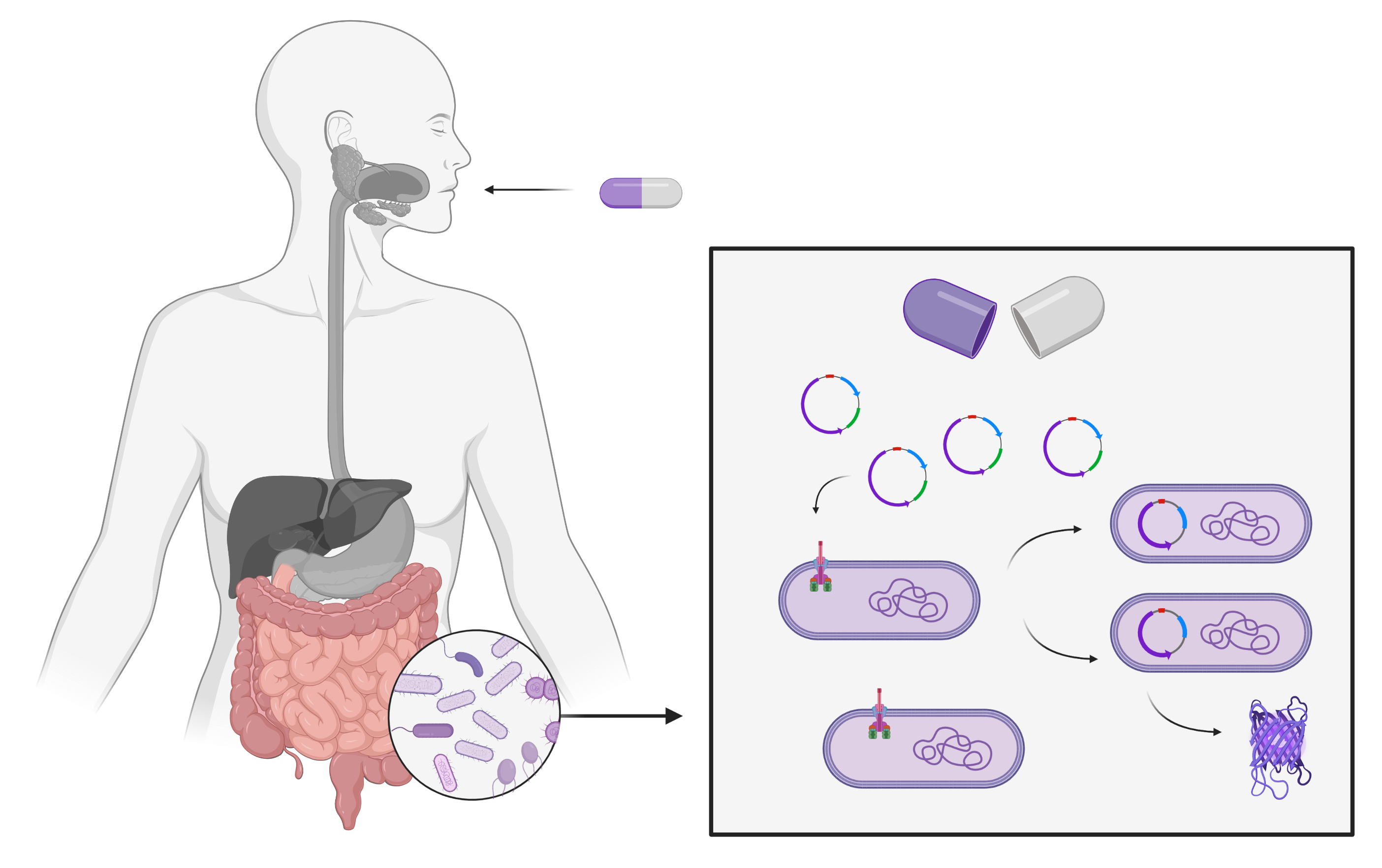
Figure 1: Transport of the transformed DNA into the gut microbiome. Created with BioRender.com
In our research project, we are focusing on the disease phenylketonuria. Phenylketonuria is a hereditary disease which causes an inability to degrade the amino acid phenylalanine. This leads to accumulations of phenylalanine in the body, impairing cognitive development during childhood. In order to treat this disease through natural transformation, we aim to modify the bacteria of the gut to express phenyl ammonia lyase as a therapeutic gene product so that phenylalanine can be degraded in the gut (Durrer et al., 2017).
Microbiome transformation: Selective advantage without antibiotics
For our project, it is important that our plasmid is not only taken up by the intestinal bacteria, but is also permanently maintained in the bacterial population. For bacteria to maintain a plasmid, it must provide them with a selective advantage. Therefore, plasmids containing an additional antibiotic resistance are often used in the laboratory. As our plasmid is supposed to be introduced into the intestine, we refrain from using an antibiotic selection. Instead, we are working on a selective advantage of the gut bacteria through dietary supplements. Basically, the additional nutritional supplements can only be metabolised by the bacteria that have taken up the plasmid, thus giving them a selective advantage. Specifically, we aim to achieve this selective advantage by introducing a beta-agarase and a resulting degradation of agar. The extent to which the plasmid is permanently maintained in the bacterial gut population, and what effects it has on it, will have to be analysed in future investigations.
Microbiome transformation: Our work in the wetlab
As many of our ideas are far too comprehensive to be fully investigated in a student project with half a year of lab work, we have concentrated on three main aspects in the laboratory.
The first one is the process of cloning by natural transformation. We confirmed that the transformation of Acinetobacter baylyi ADP1 (a strain that is known for its natural competency) works as described in the literature (Biggs et al., 2017) and further evaluated its efficiency. Moreover, we wanted to test whether the natural transformation can occur in bacterial co-cultures as well, as several bacterial strains coexist in the gut. We were able to show that the natural transformation also works when three bacterial strains, namely A. baylyi, E. coli, B. subtilis, are cultured together in the same medium (figure 2).
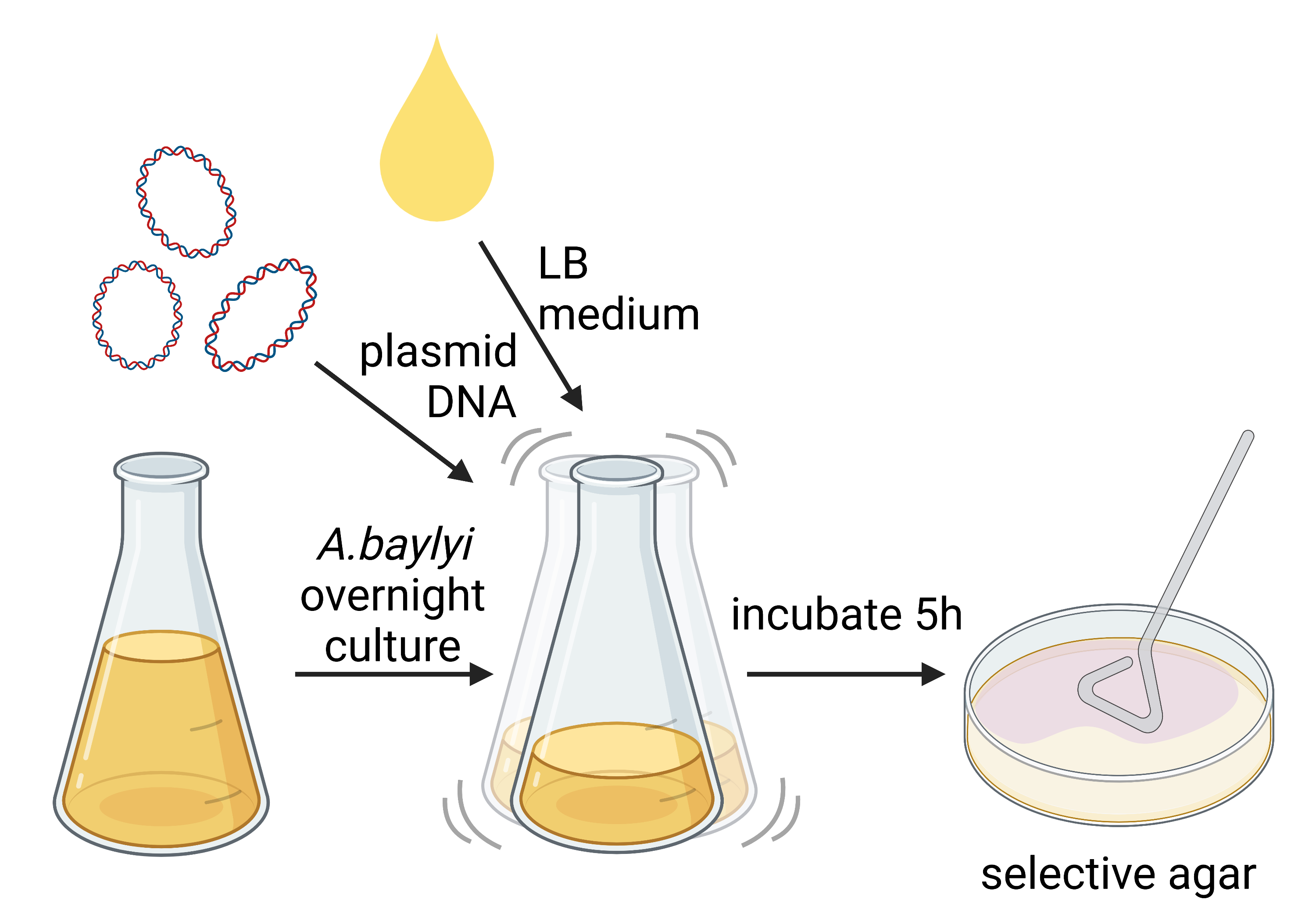
Figure 2: Depiction of natural transformation experiment. Created with BioRender.com
Secondly, we have worked on the selective advantage. We cloned the gene encoding for beta-agarase into E. coli and were able to show that the transformed E. coli degraded the agar on which it was plated. We also used the co-culture system in order to test whether the bacteria expressing the beta-agarase have an advantage over the non-transformed bacteria due to their acquired agarolytic abilities (figure 3).
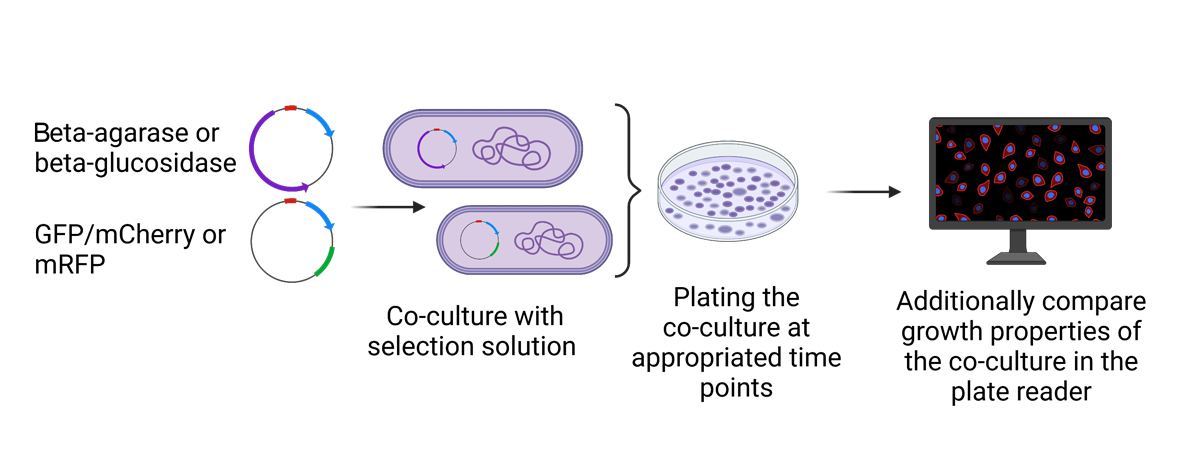
Figure 3: Depiction of the selection advantage experiment. Created with BioRender.com
The third aspect of our project focuses on our “therapeutic gene product” the phenylalanine ammonia lyase (PAL). We cloned the PAL gene into E. coli and measured its enzyme activity by detecting phenylalanine degradation. As a next step, we aim to bring PAL into A. baylyi via natural transformation.
Microbiome transformation: The problems and opportunities of the microbiome complexity
As the whole complexity of this model cannot be grasped fully and the recreation of an exact replica of the microbiome will not be possible in the wetlab, we will need to use simulations and create models based on existing data. This will be examined in the drylab, hopefully leading to meaningful findings which can then become a useful tool for future experiments in this field. The results could contribute to the prevention or treatment of more complex diseases such as obesity, inflammatory bowel disease, type II diabetes or colorectal cancer. However, there are still knowledge gaps concerning the genetic and epigenetic etiology (study of causation) of these diseases as well as the contribution of the gut microbiome to them. These knowledge gaps represent major limiting factors in designing efficient therapies.
In order to broaden our knowledge, we decided to combine genomic, transcriptomic and metagenomic data from already-published studies and utilise computational biology methods to find associations between gene expression and microbiome composition. By comparing different layers of data between healthy individuals and sufferers of disease, we hope to identify not only the genes that are upregulated or downregulated in a particular disease, but also identify microbes and microbial genes that can influence the expression of these disease-associated genes. We believe that in the future this knowledge, coupled with the technology which we are developing, can be used to design efficient therapies and in turn improve the lives of many people suffering from metabolic and gastrointestinal diseases.
The Team

Group Picture: top (left to right): Jonathan Herth, Nick Wehrwein, Franziska Gießler, Michelle Emmert, Marilena Wittmaack; bottom (left to right): Laura Sanchis, Helena Weinig, Silja Malkewitz, Kamil Moskal; additional team members: Elizaveta Bobkova, Duc Thien Bui (not shown).
By Duc Thien Bui, Franziska Gießler and Dr Andreas Ebertz
More iGEM projects:
Read about the projects of young scientists who work on solutions for current and future problems.
Did you like this article about microbiome transformation? Then subscribe to our Newsletter and we will keep you informed about our next blog posts. Subscribe to the Eurofins Genomics Newsletter.







One Comment