Sensing and converting furfural in growth media for Aspergillus niger
Lignocellulose is the most abundant biomass on Earth and is found in all plants, from trees and undergrowth in rainforests to crops on fields. Forestry and agriculture are huge industries that produce large amounts of biomass, but not all of it is used. This includes large amounts of lignocellulosic waste in the form of saw dust, rice husk and straw, of which a large part is discarded or burned to this day (Anwar et al., 2014). In the fields of farming and forestry, “sustainably available residues” is a term used to describe leftover biomass from production, of which most is lignocellulosic. France, Germany and the UK combined produced roughly 100 million tonnes sustainably available residue (Harrison et al., 2014). Some of these residues have to be left in fields to protect against soil erosion and carbon loss, while some are being used for animal feed. A large amount, however, still remains as untapped potential.
Lignocellulose (figure 1) is a complex biomass consisting of varying levels of cellulose, lignin, and hemicellulose. Due to the complexity of lignocellulose, it is difficult to efficiently utilise it as a growth media for microorganisms, thus some preprocessing is required to reduce the complexity. A commonly used method is to use a mixture of enzymes that are capable of breaking down the branches of cellulose and hemicellulose into simpler sugars such as sucrose, glucose and xylose. Lignin is often left behind and not utilised in subsequent fermentations, making it a waste product that is frequently burnt.
Due to the complexity of lignocellulose, it is difficult to efficiently utilise it in growth media for microorganisms, thus some pre-processing is required to reduce its complexity.
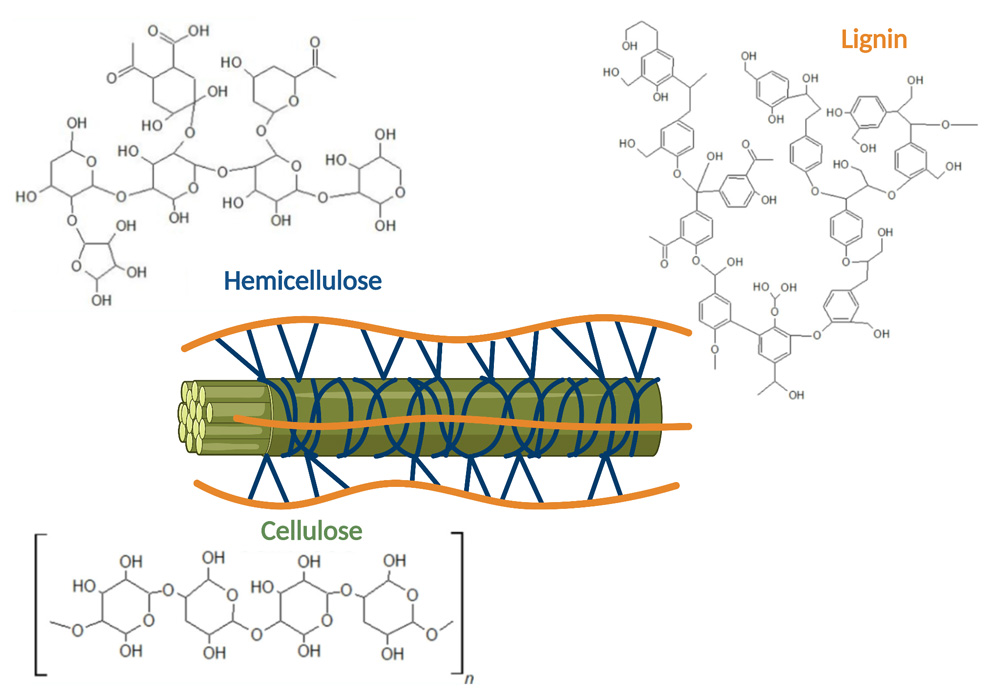
Figure 1: Illustration of lignocellulosic biomass and its three different main components cellulose (green), hemicellulose (blue) and lignin (orange) that represent the main components of plant cell walls. Figure created by using Biorender.
In order to make the cellulose and hemicellulose in the biomass easily accessible to the enzymes that convert them into simple sugars, the biomass is often processed under high pressure and heat.
Using waste products for the production of enzymes and other industrially relevant products is highly sustainable as this feeds the carbon and energy from the waste into the top of the Bio-based economy value pyramid (figure 2), allowing it to trickle down instead of being fed into the pyramid at the very bottom. Utilising waste products for the production of growth media for microorganisms is also considerably cheaper than the current production processes.
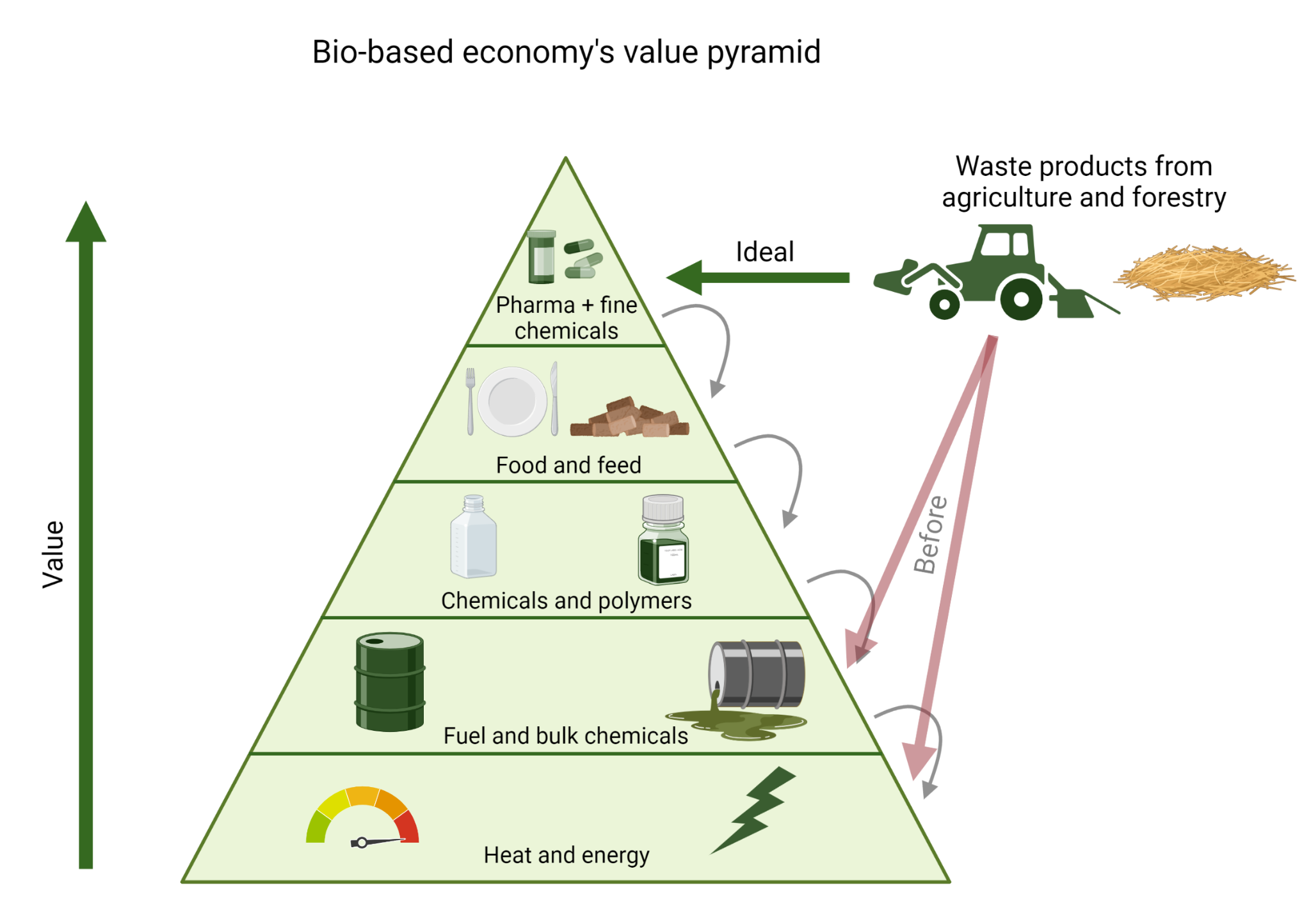
Figure 2: The Bio-based economy value pyramid. Figure created using Biorender.
However, during the pre-processing of lignocellulosic biomass, growth inhibitors are generated and released. These growth inhibitors hamper the productivity and viability of the microorganisms and thus the applicability of lignocellulosic waste products as growth media. Furfural is one of these inhibitors. Furfural has been recognised as one of the most harmful toxic compounds generated by waste processing (Heer and Sauer, 2008).
Aspergillus niger is a versatile and productive filamentous fungi, used in many industries including foods, pharmaceuticals and cosmetics. It can produce large quantities of simple compounds such as citric acid and more complex molecules such as enzymes, and is particularly well-suited for secretion of these products. The importance of citric acid in particular is shown in the global production of 2.7 million tonnes in 2021 (Imarc, 2022). A. niger has also been shown to grow well in media containing lignocellulosic waste, which is a huge benefactor for our use case, however, furfural greatly hampers the growth of A. niger even at low concentrations.
Figure 3: Cultured dish of A. niger on minimal media.
To allow for more sustainable production of A. niger when grown in lignocellulosic material, the DTU Biobuilders 2022 Team aims to genetically engineer A. niger with the goal of integrating a biosensor for furfural and genes encoding enzymes converting furfural into harmless compounds. This enables the filamentous fungi to convert furfural and thus tolerate the presence of furfural.
Detection of furfural
To our knowledge, there are no known biosensors that detect furfural, thus, we have taken several approaches to create a functional furfural biosensor from scratch. Our methods include creating a bioinformatic pipeline to find upregulated genes and their promoters in A. niger when the fungus is exposed to furfural. Without knowing the mechanisms of regulation, these promoters could in principle be used as biosensors that activate the following genes in the presence of furfural. The other method, which also adds to the novelty of our project, is the synthetic expression system (SES) using a synthetic transcription factor (sTF) (Rantasalo et. al. 2018). Our SES is composed of two transcriptional units, one unit that constitutively expresses a sTF, facilitating transcription of the other transcriptional unit, when induced by furfural. The other transcriptional unit expresses a gene of interest, in our case mCherry (figure 4).
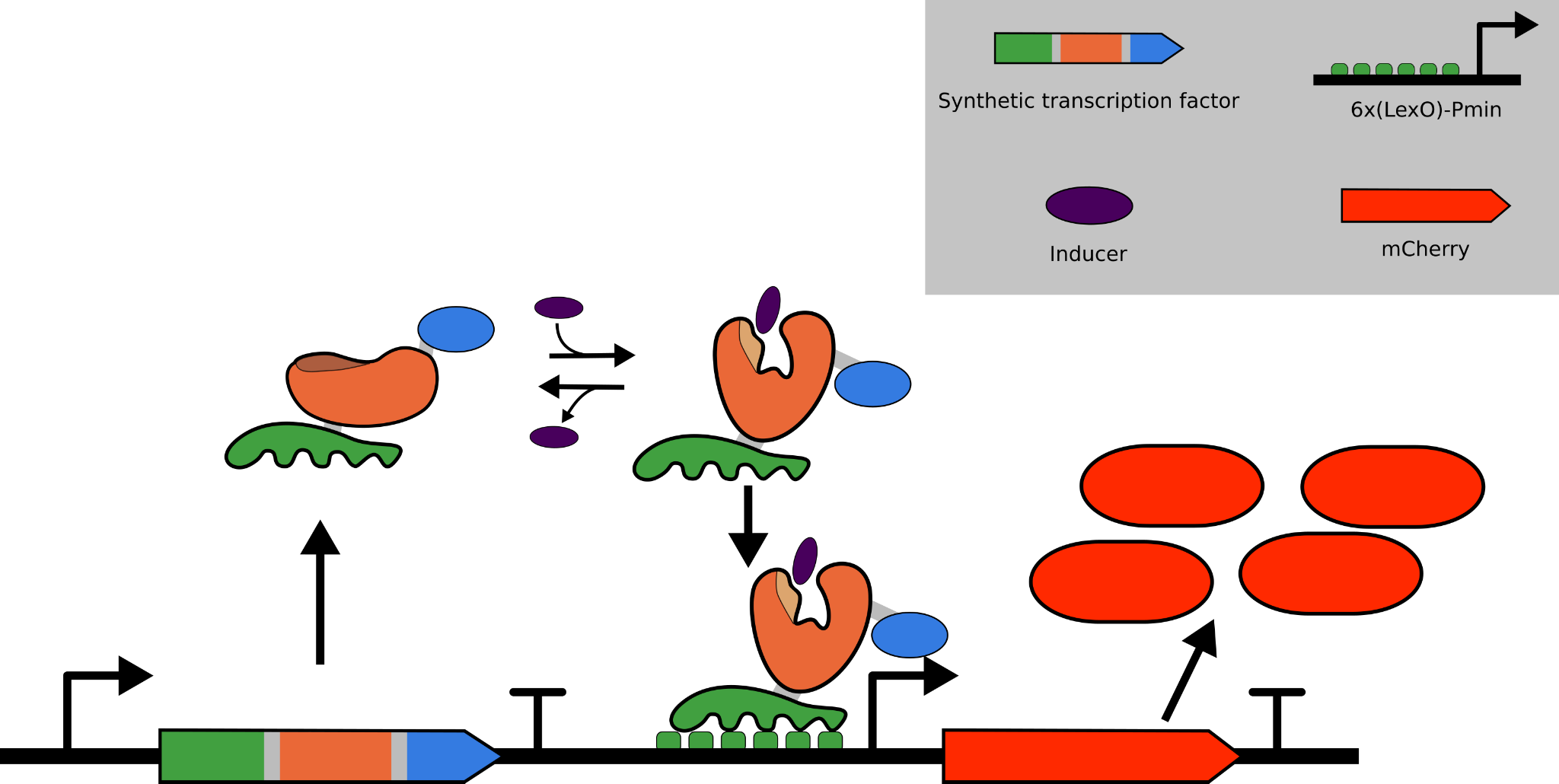
Figure 4: SES, with modular sTF gene upstream of sTF regulated promoter and mCherry.
Our synthetic transcription factors were designed to be modular, allowing for a lego-style diversification. Each module has its own functional domains and the sTF consists of three domains in total:
- DNA-binding domain (LexA)
- ligand sensing domain (HbaRe, HbaRj or Hmox1)
- transactivation domain (B112 or VP16).
The major challenge with the sTF approach is to have a ligand sensing domain that is induced by furfural. The two ligand sensing domains are HbaRe and HbaRj, where the latter is a variant of HbaRe. The domain originated from Rhodopseudomonas palustris and HbaRe has already been used in a sTF, namely a sensor of benzoic acid derivatives (sBAD) in S. cerevisiae. Hmox1 is a gene from Homo sapiens, which was computational shown to bind furfural (Santhakumar et. al. 2021).
Benzoic acid is chemically similar to furfural (figure 5) and a rational mutagenesis approach was used to increase furfural binding affinity for HbaRe and HbaRj.
Figure 5: A) Chemical structure of furfural. B) chemical structure of benzoic acid.
The different sTFs need to activate transcription of our synthetic promoter. This synthetic promoter consists of the minimal promoter from the TetOn system and six lexO binding sites. The lexO binding sites are fused in front of the minimal promoter to amplify the expression and increase specific binding of our sTFs.
Currently, the only functional SES in filamentous fungi are constitutive and not inducible. If we succeed in engineering a working inducible SES, it will be the first working case in filamentous fungi, a foundational advance allowing for specific tailoring of response and production in filamentous fungi.
Converting of furfural
A couple of furfural converting pathways have previously been discovered in different prokaryotes and fungi, such as Cupriavidus basilensis and Amorphotheca resinae. Integrating some of the converting enzymes found in these pathways into a A. niger strain should increase the productivity and viability of the strain when exposed to furfural.
The enzymes hmfH from C. basilensis and arz7774 from A. resinae, in particular, have been shown to catalyse the reaction from furfural into furoic acid, a compound that is far less toxic to cells. The conversion is a one-step reaction capable of detoxifying the growth media of A. niger when lignocellulosic waste products are used.
By integrating the genes encoding the furfural converting enzymes coupled to constitutive promoters into plasmids, we were able to test the efficiency of the enzymes. Full conversion pathways for furfural exist in both C. basilensis and A. resinae, however, it is out of scope for this project to implement a full conversion pathway within the timeframe provided by the iGEM competition.
In the future, it will be pivotal to utilise waste products to the fullest in order to minimise the carbon footprint of the human species, so there is no time to waste. Let us convert furfural in lignocellulosic media by using A. niger for a better, less wasteful, more sustainable world!
DTU Biobuilders 2022
This year’s DTU iGEM consists of 10 master’s students from DTU studying Biotechnology and Bioinformatics. We have a wide range of experience working with synthetic biology in a wide range of organisms such as bacteria, yeast, filamentous fungi, plant cells, and mammalian cells. This year we are competing in the Overgraduate category in the Biomanufacturing track.

Written by Christian Wistrøm, Dóra Katalin Jánoska, Magnus Haahr, Nanna Koefoed, Sebastian Julius Bundgaard, Trine Bertram Rasmussen and Dr. Andreas Ebertz







