The coronavirus SARS-CoV-2 and resulting disease COVID-19 have reached almost every country on Earth… but compared with other infectious diseases such as the flu, there are many unknowns about SARS-CoV-2. Scientists around the world are tirelessly researching the virus in order to understand it and eventually find treatments. What is currently known about the genomics of SARS-CoV-2 and how can the information be used in diagnostics?
What kind of virus is SARS-CoV-2?
SARS-CoV-2 is an RNA virus and belongs to the family of Coronaviridae and the genus Betacoronavirus. Its genome consists of a single-stranded positive-sense RNA. The peculiarity of single-stranded positive-sense RNA is that it can directly function as mRNA in the host cell and, therefore, is directly translated. In contrast, the RNA of negative-sense single-stranded RNA virus first needs to be converted into positive-sense RNA in order to be translated (Figure 1).
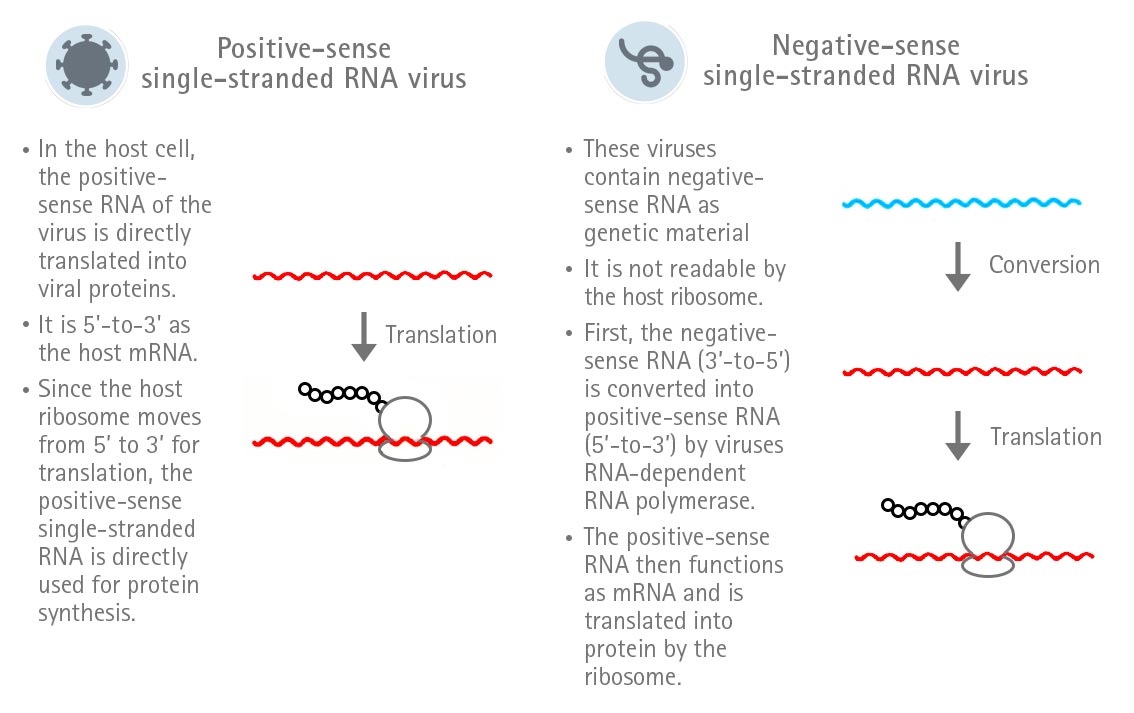
Figure 1: Expression strategies of positive-sense and negative-sense single-stranded RNA viruses.
Genomic analysis has shown that the SARS-CoV-2 genome is more similar to the genome of SARS-CoV (79% similarity) as compared to MERS-CoV (50% similarity).
Despite the genomic similarity, biological differences among these viruses are striking. The transmission rate of SARS-CoV was very low and the one of MERS-CoV was very poor at human-human transmission. In contrast, SARS-CoV-2 is the most infectious of these three coronaviruses with a transmission rate of 3.28% (Zhang and Holmes, 2020).
What is the genomic make-up of SARS-CoV-2?
The RNA genome of SARS-CoV-2 is approximately 30,000 nucleotides long and encodes 27 non-structural proteins and 4 structural proteins. The genomic composition of SARS-CoV-2 has been analysed and the sequence is publically available at the GenBank sequence repository. As of now, 104 strains of SARS-CoV-2 have been isolated and sequenced.
The 27 non-structural proteins include RNA-dependent RNA polymerase (RdRP) that was found to be highly similar to parts of the RdRP gene of coronavirus RaTG13 in bats. Other non-structural proteins are proteases and helicases for instance (Figure 2).
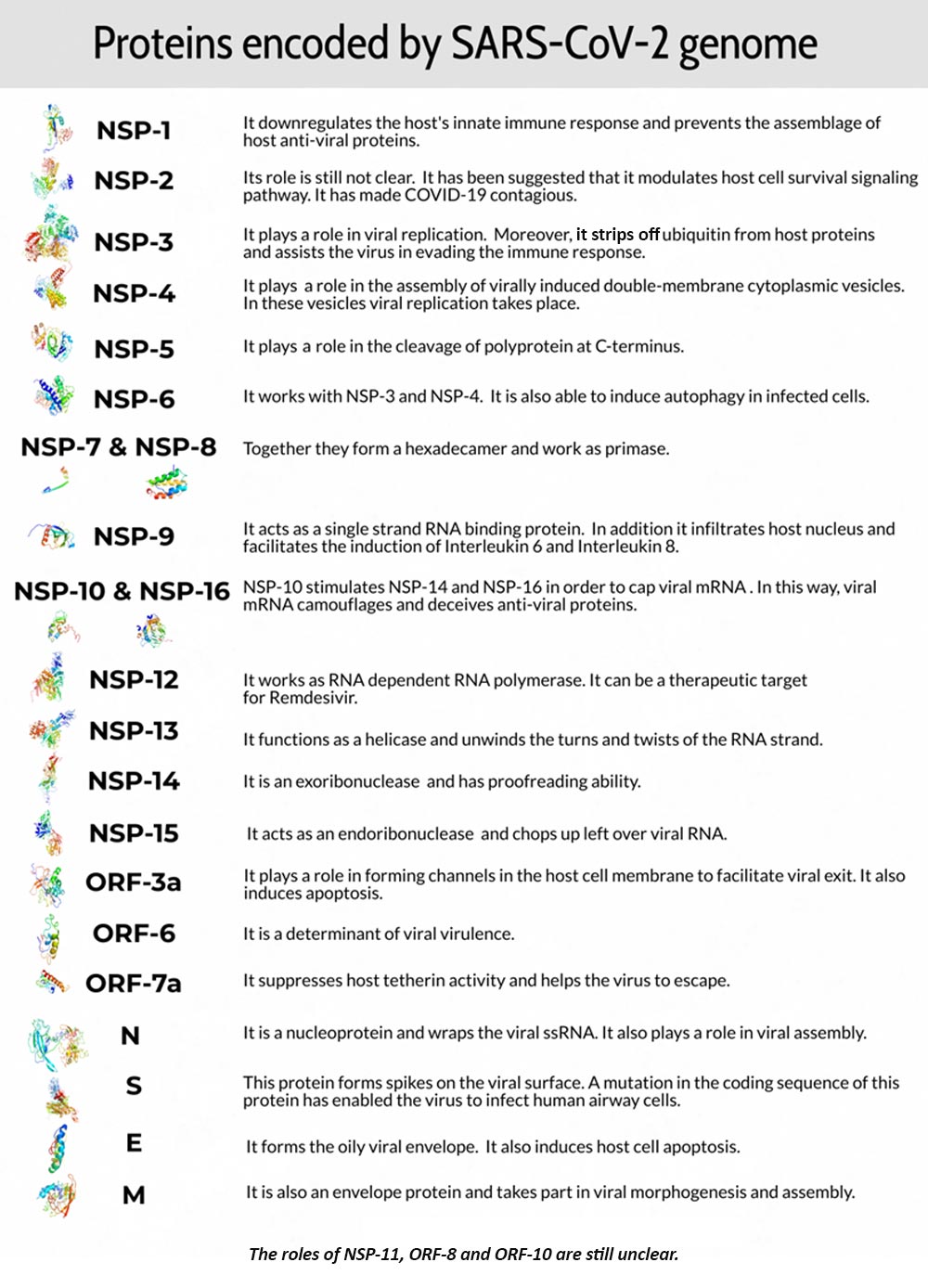
Figure 2: A summary of the SARS-CoV-2 phenotype (UCSC genome browser data; Task Force, BASE Medicine, 2020)
The 4 genes encoding structural proteins that essentially form the envelope of the virus are: 1. the spike surface glycoprotein of the virus (S), 2. matrix protein (M), 3. nucleocapsid protein (N), and 3. envelope protein (E).
Overall, the genome of SARS-CoV-2 is most similar to SARS-CoV. Genome sequence analysis has shown a 12 nucleotide insertion in the gene sequence that encodes the spike protein as compared to the SARS-CoV genome sequence that does not include this insertion. The spike proteins on the surface of the virus seem to mediate the binding of the virus to the angiotensin converting enzyme 2 (ACE2) receptors of host cells and the entry into the host cells. The spike proteins can bind strongly to ACE2 receptors due to this insertion mutation. It has been found that the binding of the spike proteins to ACE2 receptors eventually result in down-regulation of the ACE2 receptor that ultimately leads to enhanced production of angiotensin II. It has been hypothesised that this leads to lung injury. This is the reason ACE2 inhibitors can lead to critical COVID-19 disease outcomes (Diaz, 2020).
Individuals with SARS-CoV-2 present a wide variety of symptoms. Most of the patients remain asymptomatic, many have mild disease and a few patients (20%) progress to severe disease. What is the reason for this?
Can genes reveal SARS-CoV-2 mystery?
The genomic make-up of the infected individual might explain this difference why some patients only get fever and cough while others get fatigue and diarrhoea?
In the 1980s, HIV killed many people. On the other hand, HIV is not able to replicate in some individuals. These individuals carry a 32 base-pair deletion in the CCR5 receptor that renders them resistant to HIV. This mutation disables the binding of HIV to their target receptors. This could also be the case for SARS-CoV-2. Moreover, researchers are convinced that protective mutations like these may lie in genes whose expression is responsible for immune response. Michael R. Snyder, chair of the genetics department at Stanford University, has advocated this idea. “In general, we know that genetics do influence the course of a viral infection,” said Snyder. “It’s logical that immune systems are tuned differently inside different people” (Molteni, 2020).
Many universities and companies are working on genome-wide association studies (GWAS) to sift through the DNA and find out protective mutations.
Can vaccine be synthesised against SARS-CoV-2?
The world has seen the SARS (2003) and MERS (2012) epidemics and now the SARS-CoV-2 (2019) pandemic. Yet, no coronavirus vaccine has been designed to prevent humans from coronavirus-associated respiratory infections. There are two main reasons for this:
- Rapid and bulk production of a vaccine against risk group 4 organisms is challenging. Microorganisms that produce life-threatening disease are classified as risk group 4 organisms. These microorganisms pose significant risks to individuals and the community. Moreover, no treatment is available for the diseases they produce. These organisms can only be dealt with at biosafety level 3. SARS-CoV-2 is also a risk group 4 microorganism.
- In animal models, immunopotentiation in the form of lung eosinophil (specialised cell of the immune system) infiltration has been observed in the case of previous vaccines designed against MERS-CoV. Lung mononuclear infiltration was seen in all the mice models infected with MERS-CoV. In contrast, vaccinated mice that were later infected with MERS-CoV showed increased eosinophil infiltration along with an increase in IL-5 and IL-13 (eosinophil promoting chemical signals) (Agrawal et al., 2016).
A vaccine should be safe and produce strong and long-lasting immunity. The production of classical vaccines like the live attenuated vaccine and inactivated viral vaccine is under consideration. Live attenuated vaccines that are generated by utilising reverse genetics to delete virulence genes have proven to be most robust. These vaccines have the inherent ability to stimulate toll-like receptors (an immune response). At the University of Hong Kong, researchers have already developed a live attenuated vaccine (Chen et al., 2020). Here, virulence elements were deleted from the viral genome. It is a flu based vaccine that uses a flu vector to express a specific antigen.
Novel vaccine technologies are subunit vaccines, recombinant viral vectors and nucleic acid vaccines. These vaccines mimic attenuated viruses in order to generate cellular and humoral immune response. SARS-CoV-2 vaccines that are under-development include the spike protein or specifically the receptor-binding domain (RBD) of the spike protein. Nucleic acid vaccines also encode the RBD or spike protein. DNA or mRNA vaccines are safe and stable. Until now, their efficacy and immunogenicity have not been established in humans (Saif, 2020).
How is SARS-CoV-2 identified?
Molecular techniques have been shown to be highly suitable for the identification of SARS-CoV-2 infections. Specifically, real-time RT-PCR (qRT-PCR) is utilised for the analysis of respiratory samples. Here, the RNA of SARS-CoV-2 is extracted from the respiratory samples and subsequently reverse transcribed into complementary DNA (cDNA). The cDNA is then used for real-time PCR amplification with specific primers and probes that target three regions of the viral genome:
- The RdRP gene is situated in open reading frame ORF1ab.
- The E gene
- The N gene
For identification purposes, it has been suggested to use assays that target two regions. One primer pair for the identification of a broader range of coronaviruses including SARS-CoV-2, and one primer pair that is specific for SARS-CoV-2:
- The recommendation by a research group at Charité Universitätsmedizin Berlin, Germany, that is the basis of the WHO recommendation, is to first utilise a primer pair that targets different regions of the E gene to detect all SARS-related viruses. In case this leads to amplification, a second analysis with a primer pair that targets different regions of the RdRP gene is performed as confirmatory testing.
- The utilisation of a primer pair that targets the N gene and one that targets the ORF1b/RNAse P gene to confirm the results have also been proposed and are recommended by Centers for Disease Control and Prevention (CDC) (Figure 3).
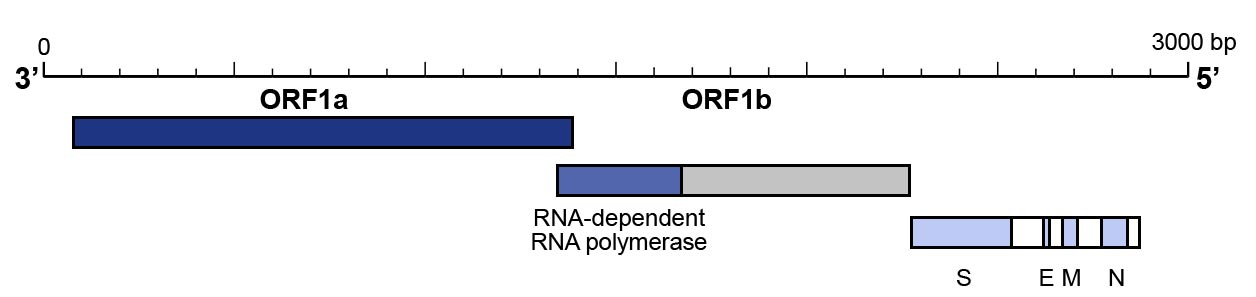
Figure 3: Genomic organisation of SARS-CoV-2
qRT-PCR can be performed in a one-step or a two-step assay. The one-step assay with reverse transcription and PCR amplification in one reaction is fast and gives reproducible results for high-throughput analysis:
- The CDC utilises a one-step qRT-PCR assay to detect a SARS-CoV-2 in samples. Here, primers and a fluorophore-quencher probe Black Hole Quencher-1 (BHQ1) and fluorescein amidite (FAM) are utilised. During amplification, fluorophore-quencher probe is cleaved, which results in the emission of a fluorescent signal.
- Charité Universitätsmedizin Berlin, Germany reported to also utilise a one-step qRT-PCR assay. Here, the probes for the assay contained Blackberry Quencher (BBQ) and 6-carboxyfluorescein (6-FAM).
The two-step assay, where reverse transcription and PCR amplification are performed in different tubes, is generally more sensitive but requires more time.
The utilisation of qRT-PCR assays requires positive controls for valid and reliable analysis.
A range of validated positive controls as plasmids that include all genes of SARS-CoV-2 as recommended by the CDC and WHO for example is also available at Eurofins Genomics.
For NGS analysis, we offer a dedicated product for full-length genome sequencing of SARS-CoV-2 based on total RNA sequencing.
By Dr. Andreas Ebertz and Tamseel Fatima





Like!! Great article post.Really thank you! Really Cool.
Thank you!
Hi Andreas,
I think you just thanked a bot.
Real question from a real person here:
What is the accuracy, false negatives and false positives, from the qRT-PCR you perform?
Hi, Yes, could be a bot.
Back to your questions:
I am not authorised to tell you how accurate the qPCR is. Sorry about that. However, we did not find virus copies in samples from healthy volunteers.
This is an excellent article because it is well-written and covers relevant information for such a time as this.
Thank you! I am glad you like it.