Oligonucleotides are a major and essential component in molecular biological research and development. In recent years, the range of applications for oligos has broadened in innovative and creative directions, such as disease diagnostics. In this article, we will show you interesting diagnostics applications of oligos.
Key takeaways:
- Oligonucleotides (oligos) are short single strands of DNA or RNA.
- Applications that use oligos, such as qPCRs, detect specific microbial pathogens in samples.
- Digital PCR (dPCR) assays for circulating cell-free cardiomyocyte DNA or miRNA could complement current tests for heart attacks.
- Analysis of miRNA expression patterns in tumour samples or liquid biopsies could aid in the diagnosis and prognosis of a wide range of cancer types.
- Oligos are used in testing for genetic alterations that are causing or are associated with neurological disorders such as spinocerebellar ataxia (SCA), epilepsy and Alzheimer’s disease.
Oligos have been critical for the work of modern-day scientists broaden our understanding of biological systems. However, academic research is only one field of application.

The significance of oligos has multiplied with their increased use in diagnostics of acute/chronic diseases and cancer for instance. The rising demand for oligos among researchers and developers is reflected in an estimated growth of the global oligo synthesis market at a Compound Annual Growth Rate of around 17 % from 2021 to 2030 (Brandessence Market Research, 2021).
Oligos and PCR
Oligonucleotides are famously used as primers for polymerase chain reaction (PCR), which was invented by Kary Mullis in the 1980s and has been used for cloning and sequencing methods ever since. A primer is typically 18-22 nucleotides long and is designed to match a specific complementary sequence (target sequence).
An important variation of the PCR that is used in various diagnostics tests is quantitative PCR (qPCR). It is used for the rapid detection of DNA sequences of cancer and pathogens, such as SARS-CoV-2, for instance.
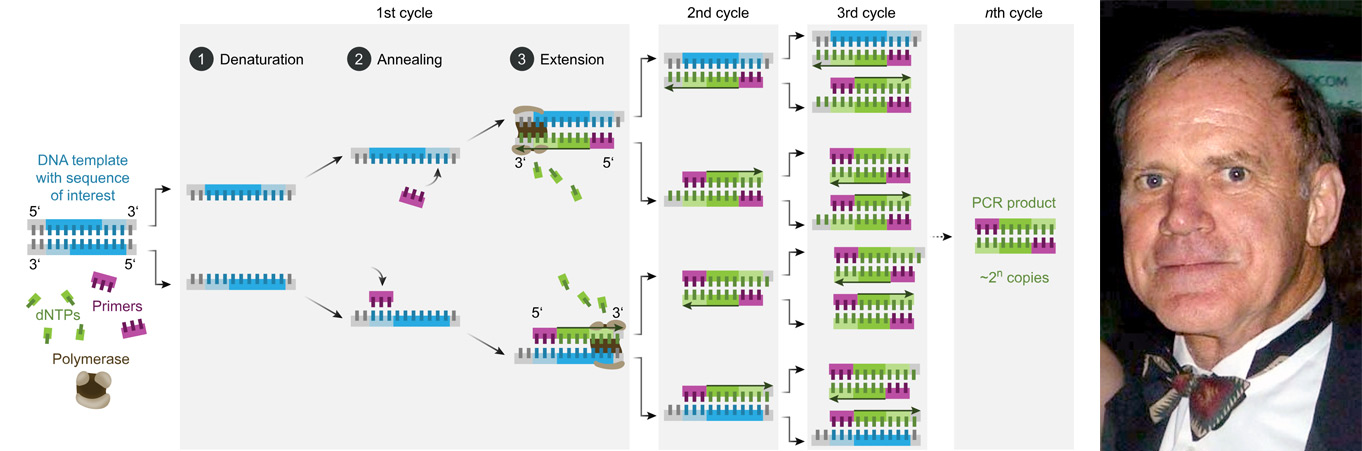
Image 1: Depiction of the PCR process and a photo of Kary Mullis, the inventor of the PCR.
In this article, you will read about different diseases and how oligos are utilised in assays and tests, such as qPCR, digital PCR (dPCR) but also microarrays and NGS, for disease diagnostics.
Oligos for diagnostics of infectious diseases
qPCR assays that utilise highly specific oligos could be an ideal tool for the rapid detection of bacterial and viral pathogens, even when only a small amount of sample material is available.
As oligonucleotides can be designed so that they only hybridise to specific pathogenic DNA or RNA, they have been used in screening assays for tuberculosis, malaria and HIV. In another application, scientists have developed assays using oligos that target pieces of Borrelia burgdorferi mRNA that encode for its toxin synthesis. This allows physicians to accurately test patients that have been bitten by ticks for Lyme disease, even if these patients do not exhibit typical symptoms associated with the tick-borne illness (image 2).
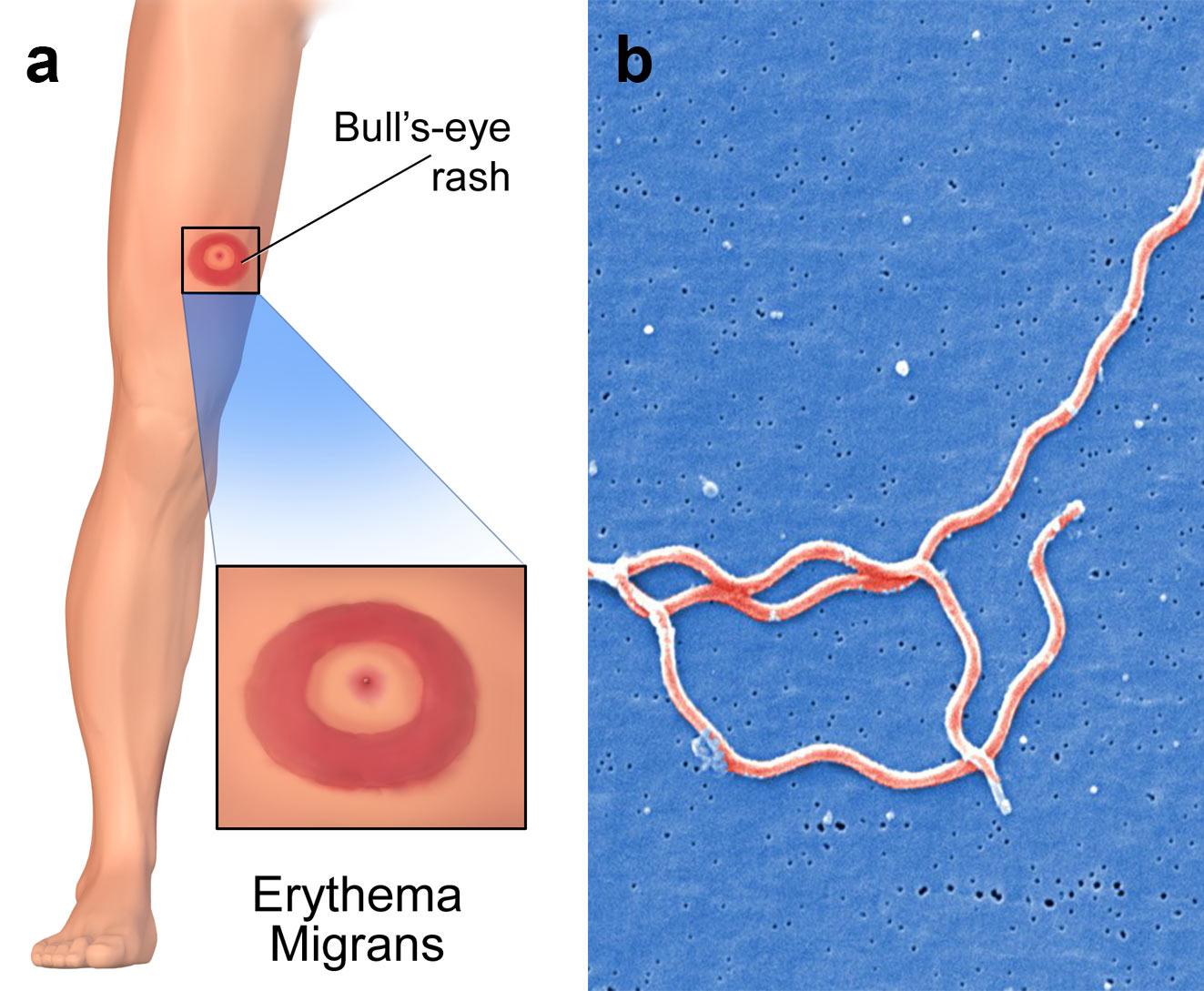
Image 2: a) Erythema migrans is the most common clinical manifestation of Lyme disease. It appears at the site of the tick bite with a mean latency of 7 to 14 days after the bite, b) three Borrelia burgdorferi bacteria.
The infrastructure in most countries infectious diseases to spread rapidly, and the emergence of new variants of concern (VOC) is much more likely. In order to identify and monitor VOC, a longer sequencing read length and high-throughput could be beneficial. Next generation sequencing (NGS) provides the ideal means for whole genome sequencing of pathogens in many samples simultaneously.
Oligos for diagnostics of cardiovascular disease
Research that utilises oligonucleotides for diagnostics of heart attacks is very promising. Assays have been developed that could complement the established tests for myocardial damage, such as for cardiac-specific troponins (cTn), which is the gold standard marker for cardiac muscle damage identification. Oligo-utilising assays could help to determine if increased cTn levels in the blood are based on cardiomyocyte death or reversible injury that can occur during taxing exercise or renal dysfunction, often found in heart disease patients.
Oligo-based assays target cardiomyocyte DNA in circulating cell-free DNA (cfDNA). As cardiomyocyte die (i.e. during a heart attack), parts of their DNA are released and enter bodily fluids, such as the blood. This cardiomycete DNA then becomes part of the cfDNA pool in the blood. A liquid biopsy, in this case a blood biopsy, is taken and screened for the presence of cardiomyocyte cfDNA; its presence would point to the occurrence of cardiomyocyte death in the patient.
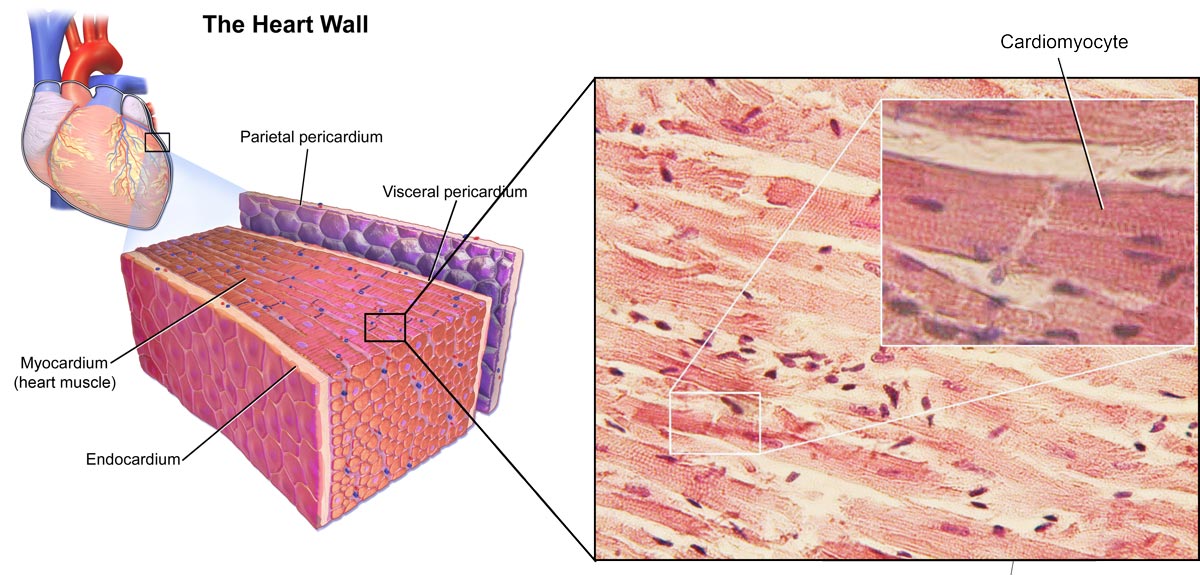
Image 3: Depiction of the heart and cardiomyocytes.
A quick return of test results is crucial to determining whether a patient complaining of chest pain is suffering a heart attack or not. Therefore, using digital PCR (dPCR) has been proposed to specifically analyse plasma samples for the presence of cardiac cfDNA in as little as seven hours after sampling.
cfDNA is DNA that is released into body fluids, such as blood, and originates from dead cells but is also secreted by living cells.
miRNAs have also been suggested as biomarkers for myocardial infarction. miRNAs are short and non-coding single stranded RNA (ssRNA) molecules that are involved in the regulation of fundamental processes such as cell proliferation, differentiation and development. miRNAs specifically bind to their target mRNAs, leading to mRNA degradation or inhibition.
Significantly, miRNA is released from the cell and can enter the blood circulation. As some miRNAs have been found to be highly tissue and disease specific, their changing levels in blood biopsies can be analysed and the results could contribute to the diagnostics of diseases. For myocardial infarction, an altered expression profile of specific miRNAs and sets of miRNAs have been identified and could act as quantifiable (via dPCR) biomarkers (Sharma et al., 2004; Catalanotto et al., 2016; Sun et al., 2017; Robinson et al., 2018; Saini et al., 2021; Zemmour et al., 2018).
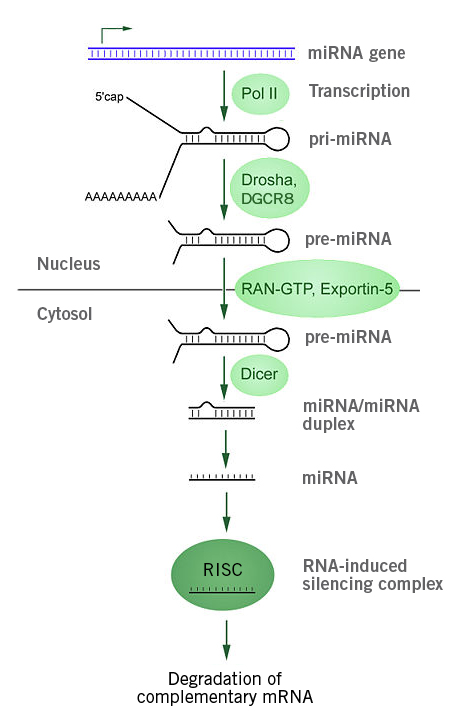
Image 4: Depiction of miRNA processing and function in animals. The miRNA gene is transcribed by RNA polymerase II into primary microRNA (pri-miRNA), which is processed by the microprocessor complex (Drosha-DGCR8) into pre-miRNAs and subsequently translocated into the cytoplasm (via exportin 5-Ran-GTP. Dicer, a RNAse III, cleaves off the loop and overhangs, producing a double stranded miRNA duplex that is used in the RNA-induced silencing complex (RISC) for post-transcriptional gene silencing. In RISC, the miRNA duplex is divided and the then mature miRNA binds to its target mRNA, leading to mRNA degradation or inhibition of translation (Image source: https://commons.wikimedia.org/wiki/File:MiRNA_processing.jpg, Created by: Narayanese, License: Creative Commons Attribution-Share Alike 3.0 Unported).
Oligos for diagnostics of cancer
Similar to myocardial infarction, the analysis of the miRNA expression patterns in liquid biopsies and solid tumour samples has been shown to be useful in the diagnostics of different types of cancers, including prostate, ovarian and breast cancers. Microarray-based studies found that tumours could be distinguished from normal tissue by their miRNA expression patterns. Even more impressively, researchers were able to classify tumours according to their tissue of origin, based on miRNA expression patterns.
In general, tumours show dysregulations of miRNA expression that have been linked to oncogenesis and affect processes such as cell proliferation, migration and apoptosis.
The global miRNA levels in various cancers (e.g. ovarian and bladder cancer) have been found to be reduced, potentially due to impaired miRNA processing in tumour cells via decreased expression and function of Drosha, Dicer 1 and DGCR8, and epigenetic regulation of miRNA. However, in cervical squamous cell carcinoma and gastric cancers, Drosha and Dicer 1 have been found to be increased, pointing to a contextual application of miRNAs as biomarkers.
In terms of highly specific miRNA biomarkers, microarray-based studies in tissue biopsies, for example, found sensitive and specific miRNAs in pancreatic cancer and mRNA/miRNA regulatory networks in colorectal cancer (CRC) that could act as diagnostic and prognostic markers, and potential therapeutic targets.
Liquid biopsies have shown great utility in cancer prediction, diagnostics and prognostics. One of the leading benefits of liquid biopsies is that they enable earlier detection of cancer compared to established tests such as CT-scan. Ultimately, early detection has the greatest impact on survival rate and time. Liquid biopsies also allow for repeated sampling to gain access to the genetic profile of the tumour over the course of the cancer treatment. The information gained could aid in therapy adjustments to improve patient outcome.
The use of miRNA-based biomarkers could add to the growing range of liquid biopsy applications. miRNAs are highly stable in bodily fluids, as they are protected from endogenous RNase activity, which makes them promising biomarkers.
The research on miRNA as biomarkers for cancer diagnostics may not have reached a point where miRNA-based assays can replace established tests, but they could complement established tests and improve overall diagnostic accuracy in the near future (Lodes et al., 2009; Wu et al., 2013; Di Leva et al., 2014; Armand-Labit and Pradines, 2017; Jiang et al., 2020).
miRNA expression analysis in (liquid) biopsies mostly utilises customised microarrays with specific oligos as probes to hybridise with the miRNAs of choice. Microarrays are ideal for discovery of new biomarkers or sample analysis in high throughput.
Biomarker validation and selection are often done by next generation sequencing, which uses oligonucleotides in the form of unique dual indices (UDIs). Find out more about biomarkers in our webinar “From Biomarker Discovery To Biomarker Validation”.
Oligos for diagnostics of neurological disorder
Neurological disorders involve the nervous system and are mainly caused by abnormalities in the brain and spinal cord.
Spinocerebellar ataxia (SCA), for example, is a group of inherited diseases (subtypes) that lead to degenerative changes in the cerebellum (part of the hindbrain, image 5) and even the spinal cord. People suffering from SCA have coordination problems, including an uncoordinated walk, accompanied by an unsteady felling or difficulty in maintaining balance.
A large percentage of the more than 40 SCA subtypes are caused by (CAG)n repeat expansions and insertions in coding exons (e.g. SCA1, SCA2, SCA3) and introns (e.g. SCA10, SCA31, SCA36).
SCA is typically diagnosed using neurological examination, CT or MRI and genetic testing. The latter is used to confirm the diagnosis and further identify the specific subtype of SCA in order to optimise treatment. The genetic tests are mostly based on microarrays or PCRs and utilise oligos as primers and probes targeting repeat expansion, among others. A wide range of providers already offer direct-to-consumer tests (ordered by a physician) (Cagnoli et al., 2018; Bird, 2019, Klockgether et al., 2019; National Ataxia Foundation, 2019).
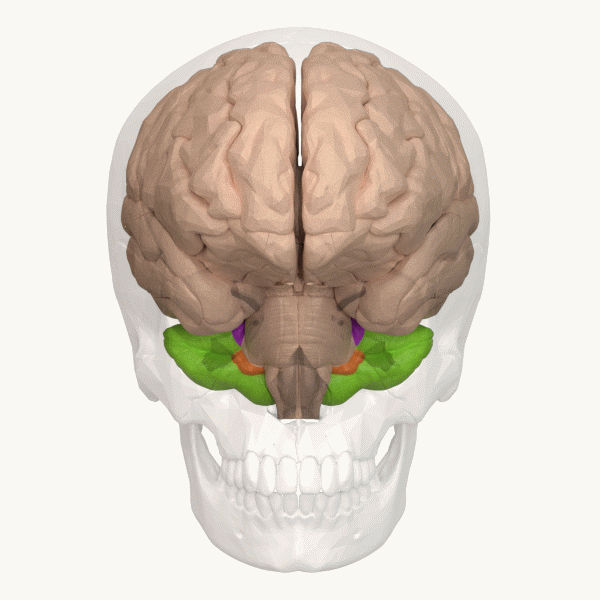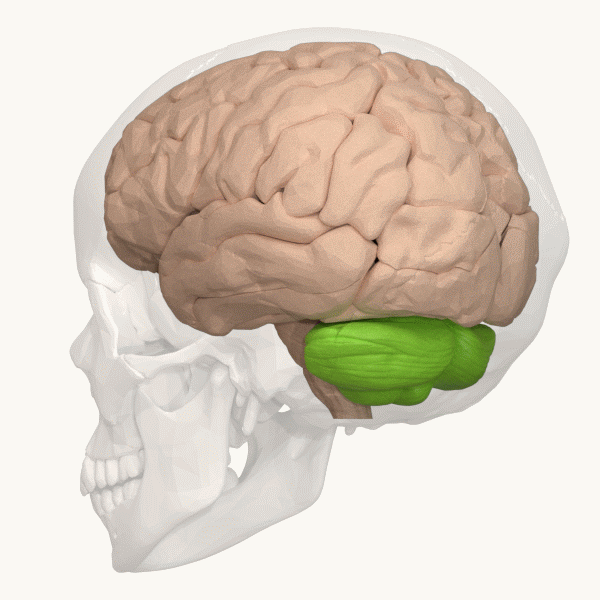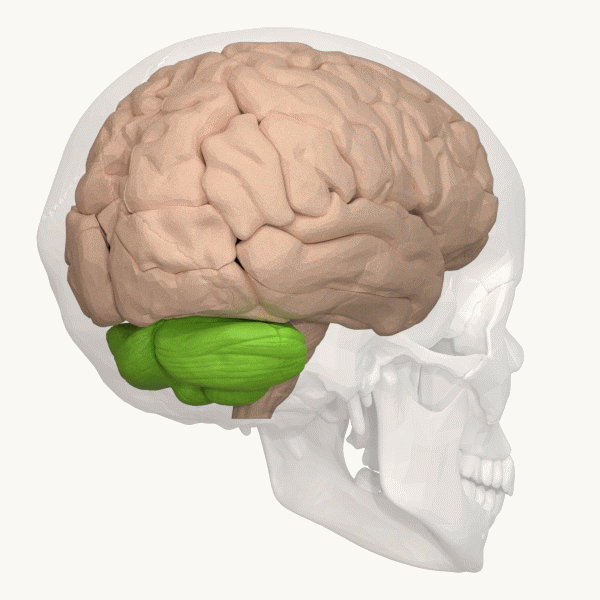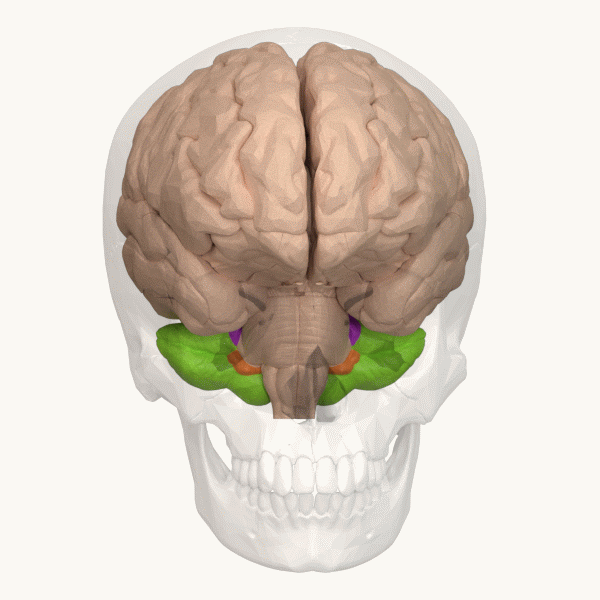
Image 6: Depiction of the cerebellum (Image source: https://commons.wikimedia.org/wiki/File:Three_lobes_of_the_cerebellum_-_animation.gif. Created by Database Center for Life Science (DBCLS); License: Creative Commons Attribution-Share Alike 2.1 Japan)
Epilepsy, another neurological disorder that affects 0.6% to 0.8% of the population can be caused by different genetic alterations. Epilepsy diagnostics has been referred to as “diagnostic odyssey” and can involve a series of investigations that could be limited by the identification of the disease-causing genetic alterations via genetic testing. The testing results could also be used to aid treatment decisions and optimisations. Similar to other diseases, genetic testing results can also be informative about the risk of family members to suffer from epilepsy.
When it comes to genetic testing, a comparative study found that targeted sequencing of epilepsy-associated genes (gene panel) and whole exome sequencing are the most cost-effective approaches. Both rely on high-throughput sequencing that utilises adapters/unique dual indexes that are forms of oligonucleotides (Poduri et al., 2014; Sánchez Fernández et al., 2019; Wojcik, 2020; McKnight et al., 2022).
Genetic tests are also available for some forms of dementia such as Alzheimer’s disease – the most common cause of dementia – and frontotemporal dementia. NGS-based tests available target panels of specific disease-associated genes in blood biopsies, buccal swabs or saliva. However, these tests are rather of predictive nature, as the utilised genes, such as APOE E4, represent risk variants. Nevertheless, tests for dementia-causing diseases that are based on mutations in single genes are available (Alzheimer’s Society, 2021; Huq et al., 2021).
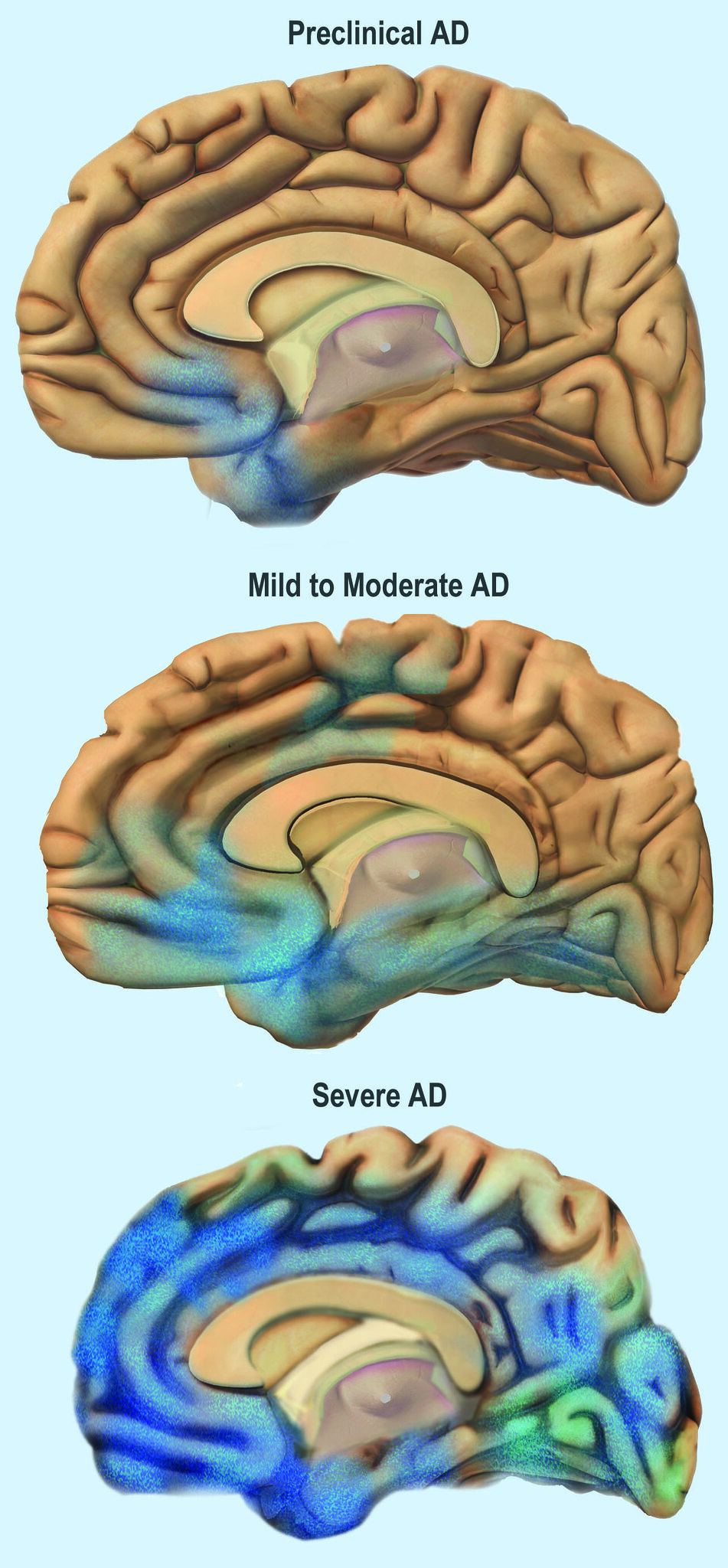
Image 7: Depiction of the progress of Alzheimer’s Disease through the brain.
The range of applications for oligonucleotides broadens into new fields and grows within existing fields. One of the fields that benefits most from the technical progress in oligo synthesis is the diagnostics market. The rising demand for genetic tests caused in parts by the increasing number of cases of genetic disorders such as cancer, as well as acceptance, requires high oligo production capacities and fast turnaround times. Find out more about qPCR Primer NightXpress, the fastest qPCR primers in Europe.
Meet the Eurofins Genomics primer synthesis expert
We are introducing Mandy Deimel, Managing Director DNA Synthesis Business. Read about her educational background, vision and get some personal insights.
Did you like this article about the use of oligos in diagnostics? Then subscribe to our Newsletter and we will keep you informed about our next blog posts. Subscribe to the Eurofins Genomics Newsletter.






3 Comments