This being said, many researchers are still not really aware of the broad range of applications for microbiome analysis; in fact that there are two main approaches.
Usual perception of microbiome analysis
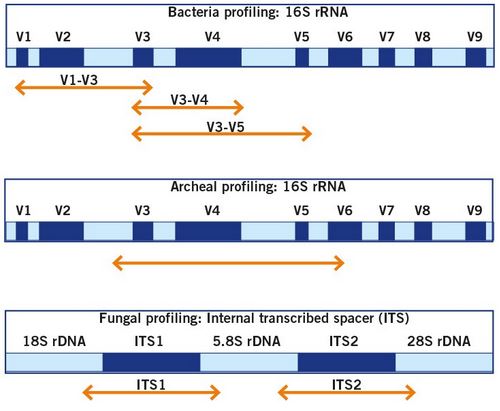
The composition of microbiomes is usually investigated by targeted amplicon sequencing that use specific genetic markers, such as variable regions of the 16S rRNA gene for bacteria and archaea, and the ITS region for fungi. This approach is widely considered as the gold standard for analysis of whole microbial communities. Here, the DNA of microorganisms is recovered from the samples and then specific marker gene regions are amplified by PCR. This mainly allows for the taxonomic classification and analysis of richness and abundances of microbial taxa in a sample.
With more than 100,000 sequences of the 16S rRNA gene available in public databases, 16S rDNA amplicon sequencing can profile hundreds of microorganisms, including low-abundance and non-cultivable bacteria from a single analysis, and can semi-quantitatively describe bacteria present in complex biological samples. This can be used to process environmental and clinical samples and serve various purposes from food quality assurance to gut microbiota research. Read more about the influence of the microbiome on human health and your options of microbiome profiling.
Enter metagenome analysis
Metagenome analysis, on the other hand, utilise the entire genetic content of all microorganisms for analysis by sequencing the whole genome. Here, the whole microbial DNA in a sample is recovered and directly used for DNA sequencing. This also allows for the taxonomic classification of the microorganisms, potentially to an even higher degree than the 16S rDNA amplicon sequencing approach, but it also provide insights into the function of genes and gene clusters such as enzymes and biocatalysts, and into genomic linkages between community function and structure.
This is best illustrated and explained by examples for applications of metagenome analysis.
Metagenome analysis in cancer research
It has been estimated that more than 15% of the global cancer burden is linked to microbes. Metagenome analysis could serve as an approach to identify and analyse the microbial diversity present in cancer tissue specimens. By generating a comprehensive metagenomic view of cancer microbiota, linkages between microorganisms and various types of cancers could be established. The identification of potential microorganisms associated with a particular cancer could lead to greatly enhanced drug target development and selection.
Read more about the microbiome of cancer.
Metagenome analysis and the human gut microbiome
It has been estimated that up to 1,000 different microbial species live in the human gastro-intestinal tract. Metagenome analysis has contributed to investigations of the intestinal microbial diversity and the dysbiosis of the microbiota, as well as of the relationship of the gut microbiota with health and disease.
Functional metagenome analysis could contribute to the identification of novel functional genes, microbial pathways and antibiotic resistance genes, as well as the analysis of the co-evolution between microbiota and host. Overall, it could aid the management of human diseases.
Viral metagenome analysis
Viruses are the most abundant biological entities on the planet. Since there is no single gene that is common to all viral genomes, viruses cannot be investigated by commonly used methods such as amplicon sequencing. A collection of viral metagenomic studies showed that 60 to 99% of generated sequences are not homologous to known viruses. This underlies the importance of metagenome analysis in the field of virology. Specifically, viral metagenome analysis has provided the means for virus discovery, characterisation of normal viral population in a healthy community, and identification of viruses that could pose a threat to human health.
Metagenome analysis in the food and pharmaceutical industries
Functional metagenome analysis has considerable potential for the food and pharma industries. It could support the identification of enzymes with valuable technological properties and with the ability to function under extreme conditions. Metagenome sequencing could also be used to discover novel bioactive compounds, such as antimicrobials, and to address antibiotic resistance development. For quality control of food and medicines, the metagenome analysis approach could be a valuable tool for the detection of proper ingredients, contamination, allergens, or spoilage in food.
Environmental metagenome analysis
Metagenome analysis of microbial communities in the environment could provide valuable information for the development of biofuels, environmental remediation, discovery of agrochemicals, and improvement of agriculture practices. It can be applied to a wide range of samples, from air debris to soil samples. It has been shown that in soil samples, the microbial diversity is greater than in any other microbial ecosystem. This implies that novel microbial resources including microbial genes encoding novel enzymes or bioactive compounds could be uncovered here. Metagenome analysis is the perfect tool to aid this search.
Furthermore, metagenome sequencing could play a significant role in quality control of drinking water but also bodies of water.

Listen to the interview with the founder of Blue Biolabs, where we talk about the application of metagenome analysis for quality control of drinking water, detection of pathogens and much more.

The Eurofins Genomics’ INVIEW Metagenome service offers next-generation sequencing of the full genomes of all bacteria, archaea and virus in a given sample.

The Eurofins Genomics INVIEW Microbiome services offers next-generation sequencing of the 16S rRNA gene to enable simultaneous identification of hundreds of different bacteria, fungi and archaea without any cultivation steps.
Are you still unsure which approach to pick for your scientific question and investigation? Try our next-generation sequencing decision tree for non-human samples to find the best fitting service.
In case the depiction of the decision tree does not work properly in Internet Explorer or Firefox, please use Google Chrome.
By Dr Andreas Ebertz
Did you like this article? Then subscribe to our Newsletter and we will keep you informed about our next blog posts. Subscribe to the Eurofins Genomics Newsletter.





3 Comments