Image: Methicillin-resistant Staphylococcus aureus (MRSA) and a dead Human neutrophil
Effects of the Gut Microbiome on Digestive Health
The consistently advancing research on microbiota found fascinating interactions between the human gut microbiota and the host. First and foremost, the gut microbiota is of paramount importance for the digestion of food. One field of intense research, therefore, aims at the impact of this microbiota on nutrition and digestion-related diseases. In individuals that suffer from irritable bowel syndrome, inflammatory bowel disease, different forms of liver disease, colorectal cancer, or who develop obesity and type 2 diabetes, an altered gut microbiota has been found, compared to healthy and lean individuals.
Corrective interventions such as probiotics administration to improve different liver injuries, or faecal matter transfer from lean individuals into obese individuals with metabolic syndrome, resulted in improved health conditions. Despite early successes, a clear approach to treat these condition has not been established yet (Ridaura et al., 213; Khan et al. 2014; Dinan et al., 2015; Marchesi et al., 2015; Carding et al., 2015).
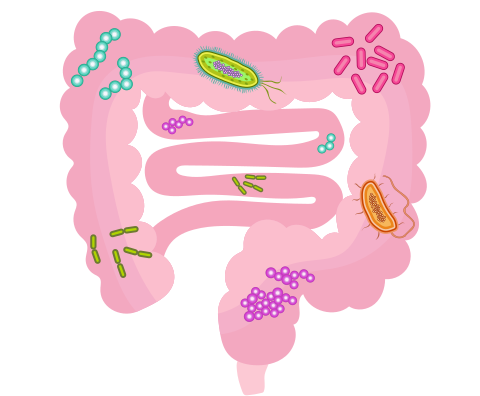
Effect of the Gut Microbiome on Metabolism, Immune System and Inflammation
The human gut microbiota, however, has much greater importance. Research indicates that it affects the host’s metabolism, immune system and inflammatory response. It is also involved in de novo synthesises essential vitamins, and even influences the host’s stress response (Artis, 2008; Dinan et al., 2015; Thursby and Juge, 2017; Marietta et al., 2017). Alteration of the gut microbiota, termed dysbiosis, has been associated with systemic diseases such as multiple sclerosis and rheumatoid arthritis, as well as autism (autism spectrum disorder) and depression (Rosser and Mauri, 2016; Cekanaviciute et. al., 2017; Luna et al., 2017; MacQueen et al., 2017). In an interspecies study, the stool samples of human MS patients were transplanted into mice and the animals showed disease symptoms similar to MS in humans, but not when stool samples of healthy patients were transplanted (Cekanaviciute et. al., 2017).
It has even been postulated that the gut microbiota could be viewed as an organ and as a therapeutic target for a more specific personalised healthcare (Marchesi et al., 2015).
Microbiota and Skin Conditions
Microbiota profiling also plays an important role in clinical testing of skin for instance. Skin diseases often affect specific areas, such as eczema (atopic dermatitis) that causes rashes on arms and legs. Certain bacterial taxa have been found in association with eczema, which raises the question of their particular involvement in this skin condition. It has been proposed that microbiome-based treatments could aid to ameliorate skin diseases (Waldor et al., 2015).
Microbiome-based therapeutic treatments are still at their beginning, and investigations with highly sensitive and flexible microbiome profiling service, as provided by the Eurofins Genomics Microbiome Profiling, will be required to broaden our knowledge on the effects of the microbiome.
Eurofins’ New Microbiome Profiling Service
Eurofins’ new Microbiome Profiling Service is a powerful end-to-end service for in-depth characterisation and bioinformatics analysis of microbial communities of any complexity. The new version provides our customers a comprehensive characterisation of the bacterial and fungal communities in their samples at an even lower price than before. You can make use of validated primers from high-impact publications for PCR amplification that cover the variable regions V1-V3, V3-V4, and V3-V5 of the bacterial 16S rRNA gene, and the ITS1 and ITS2 regions of the fungal rRNA gene.
For a better discrimination between microbe-specific amplicons, you can now choose up to 4 targets per sample, in any combination you prefer. The use of multiple primer combinations is recommended to capture the entire community, since each 16S and ITS primer combination is suited for certain taxa and less suited for other (Ihrmark et al., 2012; Toju et al., 2012).
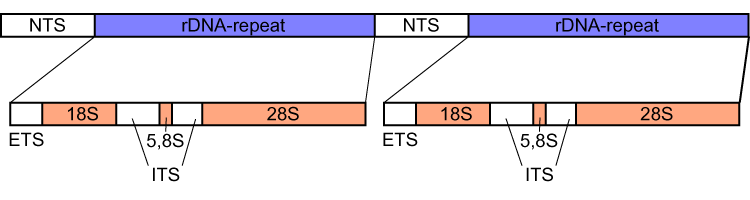
Image: Eukaryotic rDNA (NTS – nontranscribed spacer, ETS – external transcribed spacer, ITS – internal transcribed spacers)
In case you prefer your own target specific amplicons, choose Eurofins’ 2nd PCR Microbiome Profiling. We will purify and high-throughput sequence your own target-specific PCR amplicons. This option gives you even more flexibility for your experiments and investigations. It allows you to investigate the archeal community for instance, or allows you to utilise the fungal 18S rRNA gene for taxonomic identification rather than the ITS regions.
Read more about the effects of the microbiome on human health.
How are faecal transplants used for the treatment of antibiotics-associated disease?

Did you know that the microbiome profoundly affects cancer and cancer development?
Metagenome analysis – Beyond microbiome analysis
By Dr Andreas Ebertz






5 Comments