Why did we include archaea?
The current knowledge about the archaea’s detailed physiological or clinical relevance in humans is rather low but gains more and more interest, especially now since certain taxa have been found in association with conditions such as irritable bowel disease, obesity, anorexia nervosa, but also with infectious diseases like brain abscesses.It was believed that archaea are not pathogenic to humans per se, but these findings point to opportunism and potential pathogenicity (Nkamga et al., 2017).
Furthermore, research suggests diverse and important roles of archaea in the microbiomes of the human skin and mouth, growth and health promoting effects on plants, and their presence in the microbiome of corals, intracellular niches in amoebae, protozoa, and termites for instance (van Bruggen et al., 1985; Wegley et al., 2004; Chaban et al., 2006; Janssen et al., 2008; Lins-de-Barros et al., 2010; Horz and Conrads, 2011; Koskinen et al., 2017; Taffner et al., 2018).
Methanogenic archaea in the digestive tract of ruminants are the main producers of methane. Investigations of these archaea could potentially lead to reduced methane emission and a substantial long-term impact on the environment. After all, methane is an abundant greenhouse gas that impacts the greenhouse effect 25 times more than carbon dioxide (Dridi et al., 2011).
By adding the option to profile archaea to the Microbiome Profiling Service 3.0, we want to provide a tool to broaden the knowledge about the effects and impact of archaea. You can choose 1 to 4 targets per sample, 16S targets for archaea and bacteria, and ITS targets for fungi. That way you can either focus on archaea, bacteria or fungi only, or analyse the entire microbial community when choosing more targets.
One Example: The Gut Archaeome
Archaea as members of the gut microbiome and their impact on human health have been neglected in the past but are given more attention in recent research. Currently, about 20 different archaeal taxa have been found as the part of the gut microbiome of humans (Nkamga et al., 2017). Here, anaerobic methanogenic archaea constantly utilise and, thereby, remove hydrogen (H2) that is generated during bacterial fermentation processes to metabolise their substrate to methane. This seems to be of utmost importance, since it prevents the accumulation of acids and reaction end-products and contributes to the reduction of the gas partial pressure in the colon (removal of H2 and CO2) (Dridi et al., 2011; Gaci et al., 2014; Koskinen et al., 2017; Nkamga et al., 2017).
Overall, methanogenic archaea account for approximately 10 % of the anaerobic gut microbes. Sequencing-based investigations found that Methanobrevibacter smithii is the predominant archaeal species in the human gut. It can be found in more than 50 % of the adult population and, here, it represents the vast majority of archaeal species of the gut microbiome.
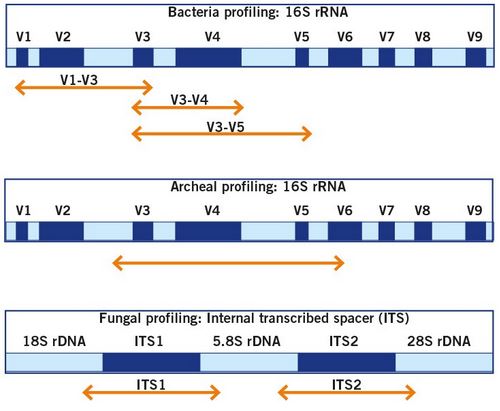
Find out how microbiome analysis is done and what the genetic targets for the identification of bacteria, archaea and fungi are.
By Dr Andreas Ebertz






2 Comments