The use of mRNA in biology
In 1953, after Watson and Crick proposed the model of the DNA double helix, a new question arose in the scientific community: how is information encoded by the DNA and how is it translated? The earliest hypothesis of the involvement of RNA came from André Boivin. He expressed his idea in a paper published in Experientia, stating “the macromolecular deoxyribonucleic acids govern the building of macro-molecular ribonucleic acids, and, in turn, these control the production of cytoplasmic enzymes.” However, during the mid-1950s not enough knowledge was available to support this hypothesis (Cobb, 2015).
Update
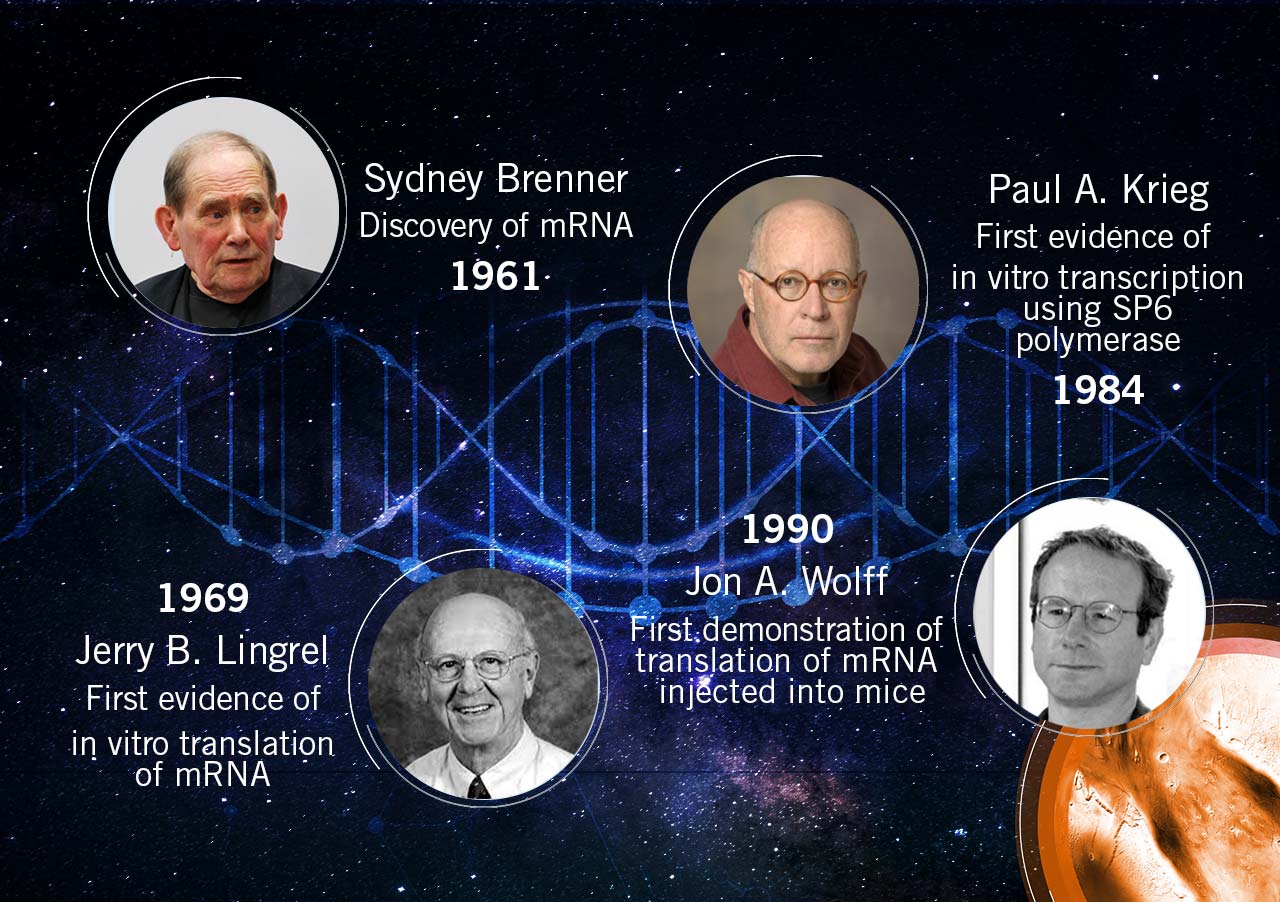
Discovery of mRNA: It started in 1961, when Brenner and colleagues described the presence of an unstable intermediate molecule that copies the information encoded by the DNA and directs the synthesis of proteins: RNA. The group around Brenner worked with virus-infected cells and analysed the gene expression. They concluded that the protein-encoding information is not present in stable ribosomal RNA. Instead, a transient RNA molecule acts as a transcript of the genetic code. This RNA was termed messenger RNA (mRNA). Ribosomes synthesise proteins according to the information dictated by mRNA (Brenner et al., 1961).
Nucleic acids can induce interferon production: In 1963, Isaacs et al. demonstrated that viral nucleic acids can induce the production of interferon in infected chick, rabbit, and mouse cells. The production of interferon supports the fact that nucleic acid is considered foreign to the cell (Isaacs et al., 1963).
In-vitro mRNA translation: In 1969, mRNA was translated in the lab for the first time. Lockard and Lingrel, who worked together at the University of Cincinnati, Ohio, provided the first evidence of in-vitro translation of mRNA. They used a mammalian (rabbit) cell-free system to demonstrate the translation of an mRNA transcript from another mammalian species (Lockard and Lingrel, 1969).
SP6 and T7 RNA polymerase: Moreover, in 1984, experimental work showed that any desired cDNA can be utilised for the synthesis of functional mRNAs and, consequently, the synthesis of proteins (Krieg and Melton, 1984). In light of this work, SP6 RNA polymerase was eventually commercialised. In addition to SP6 polymerase, researchers extracted and purified T7 RNA polymerase. In 1985, they also designed a T7 polymerase-promoter complex for the controlled expression of specific genes that is still used today (in upgraded form) (Tabor & Richardson, 1985). These pioneer experiments lead to an unstoppable series of practical work concerning mRNA delivery and commercialisation.
Liposomes as mRNA delivery vehicle: In 1978, liposomes were utilised for the delivery of mRNA to eukaryotic cells (Dimitriadis, 1978). By the end of the following decade, a cationic liposome mRNA delivery system, DOTMA, was described and commercialised (Malone et al., 1989).
The rise: mRNA as a therapeutic agent
mRNA expression in-vivo: The foundation of the concept of mRNA as a therapeutic agent was laid by Wolff J. and colleagues in 1990. The team injected naked RNA into mice muscles to provide proof of principle for direct gene transfer in vivo (Wolff et al., 1990).
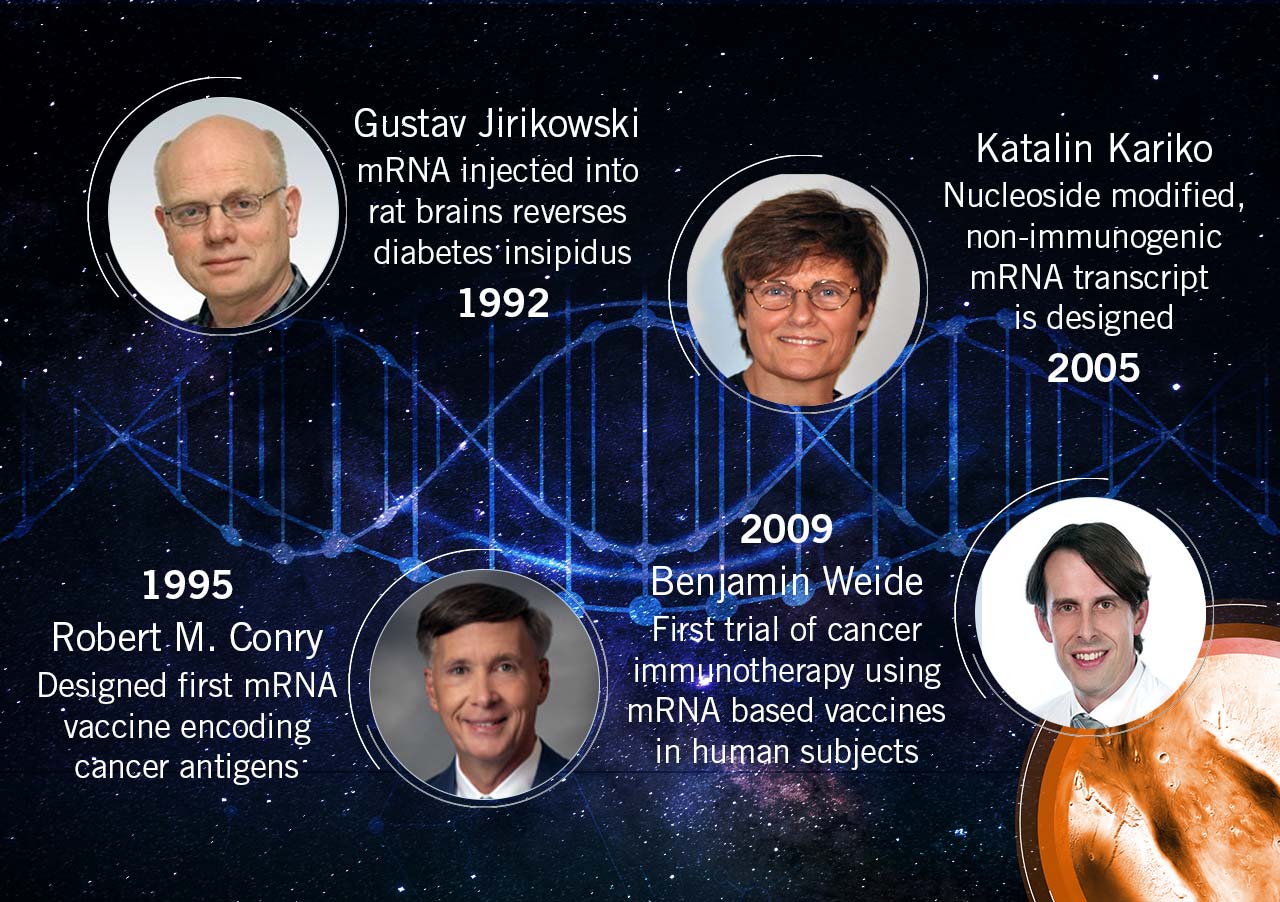
1st use of mRNA as therapeutic molecule: In 1992, a team of scientists working at Scripps Research Institute used mRNA to transiently reverse diabetes insipidus in Brattleboro rats that do not produce the hormone vasopressin (Jirikowski et al., 1992).
1st use of mRNA as vaccines: Even though the concept of mRNA vaccines sounds relatively advanced, it dates back to 1995, when Robert and his team designed the first mRNA vaccine that encoded cancer antigens (Conry et al., 1995).
1st mRNA company: All this work in mRNA therapeutics laid the cornerstone of the first mRNA company ever founded: Merix Bioscience (1997). In 2004, the company changed its name to Argos Therapeutics as a sign of its evolution.
The climax: Challenges vs solutions
The idea of using mRNA as a therapeutic agent sounds straightforward, but it is not. Two major challenges had to be overcome: 1) Successful delivery of mRNA, and 2) Prevention of an immune response against the mRNA molecule.
Isolation of RNA is difficult as ever present RNase easily degrades RNA. In order to prevent this degradation, different measures to inhibit RNase function have to be taken when RNA is isolated from cells/tissue.
Enhancing mRNA stability: When mRNA is injected into the body it triggers an immune response. As a result, the injected mRNA is degraded and the synthesis of the desired proteins is shut down.
For these reasons, little funding was granted to studies focusing on mRNA based therapeutics. Even Nobel laureate Philip A. Sharp found it to be a concept with little practicality for the above mentioned reasons. Katalin Karikó, a researcher at the University of Pennsylvania, struggled to get funds for her research on mRNA application. “I thought of going somewhere else, or doing something else,” Karikó said. “I also thought maybe I’m not good enough, not smart enough. I tried to imagine: Everything is here, and I just have to do better experiments.” Even after all the setbacks, she continued her search for that one better experiment (Leah Asmelash and AJ Willingham, 2020).
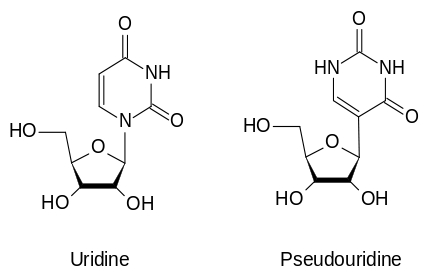
In 2015, Katalin Karikó and her colleague Drew Weissmann found the solution to prevent activation of the immune response against the injected mRNA. It has been found that mRNA activates the toll-like receptors (TLR) on immune cells. Karikó and Weissman modified the RNA by inserting a naturally occurring modified nucleoside: pseudouridine. This modified RNA transcript does not activate the TLR-mediated immune response and even provides a higher translational capacity (Karikó et al., 2005). A milestone for mRNA based therapeutics was reached. Even Sharp, the former critic, eventually said about the work done at Moderna: “They’ve totally convinced me it is possible to do”.
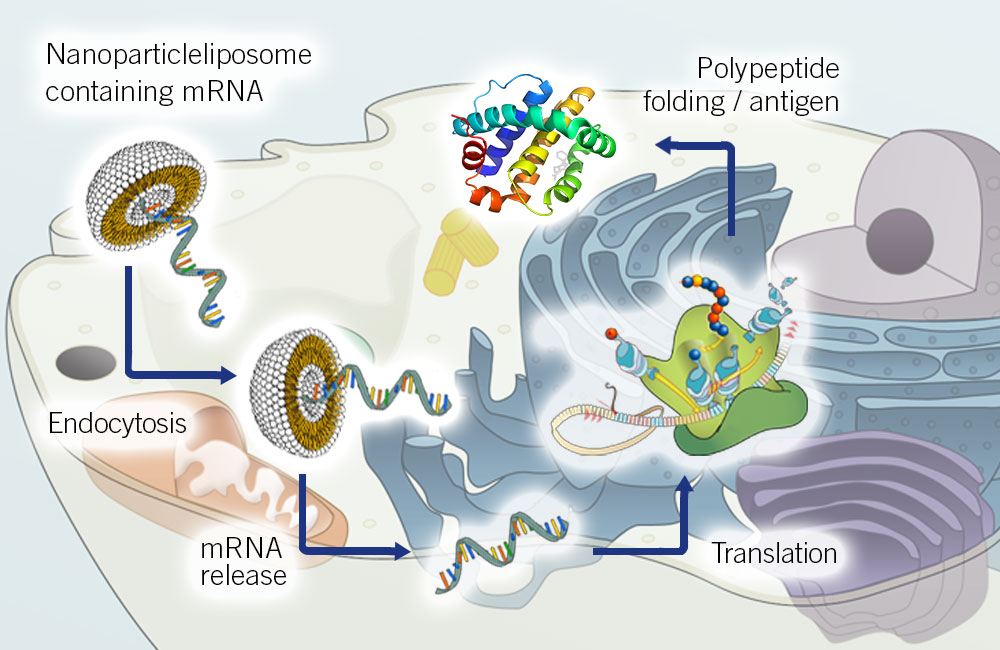
Improving RNA delivery: In parallel, Pieter Cullis at the University of British Columbia was studying lipids. In 1995, Cullis and his team started working on the application of lipid nanoparticles (liposomes) for the delivery of large biological molecules such as RNA. Even though the system did not work consistently, their work advanced the research on using lipids as an effective and safe delivery system (Bailey & Cullis, 1997).
Derrick Rossi, working at Harvard Medical School, built upon Shinya Yamanaka’s work of inducing pluripotency and used RNA to create embryonic stem cells from adult cells (Warren et al., 2010). This eventually led to the foundation of Moderna in 2010. The same year, Cullis, Karikó, and Weissman collaborated to formulate a vaccine consisting of mRNA and lipid nanoparticles. Even Sharp, the former critic, eventually said about the work done at Moderna: “They’ve totally convinced me it is possible to do.”
In-vitro transcription of mRNA: Nucleic acids have revolutionised the field of genetics and regenerative medicine. The in-vitro transcription of mRNA has addressed almost all the challenges associated with the use of mRNA as a therapeutic agent. In-vitro transcription is simple, rapid, and controlled. Most importantly, chemical modifications can be introduced to the mRNA strand in-vitro (Kwon et al., 2018).
Applications of mRNA technology
During the following years, various pre-clinical and clinical trials on mRNA-based vaccines against infectious diseases, hypersensitivities and cancer were completed (Sahin et al., 2014; Weissman, 2015).
Vaccines
In 2009, researchers conducted the first-ever trial on cancer immunotherapy using mRNA-based vaccines in human subjects with metastatic melanoma. The results of the trial showed an increase in the number of vaccine-directed T cells against melanoma (Weide et al., 2009).
In 2020, the FDA approved the first mRNA-based vaccines against an infectious disease SARS-CoV-2. This was only made possible by decades of research on mRNA-based therapeutics.
Regenerative medicine
mRNA has not only been a subject of interest for vaccine development, it has also influenced other fields including stem cell science and protein replacement therapies. Induced pluripotent stem cells (IPSCs) are a highly interesting addition to the tool-box of regenerative medicine, as IPSCs can differentiate into every other cell type in the body. The Japanese scientist Shinya Yamanaka transfected somatic cells by introducing several transcription factors and converted them to an embryonic stem cell state. That’s the dream of regenerative medicine. However, this process has the inherited danger of DNA integration into random sites of the genome and thereby potentially leading to adverse mutations, and unpredictable results. To counteract this issue, Yakubov et al. reprogrammed fibroblasts (a kind of skin cells) into IPSCs using mRNA transfection. This method completely avoids DNA integration and could be further developed to replace already available methods to generate IPSCs (Yakubov et al., 2010).
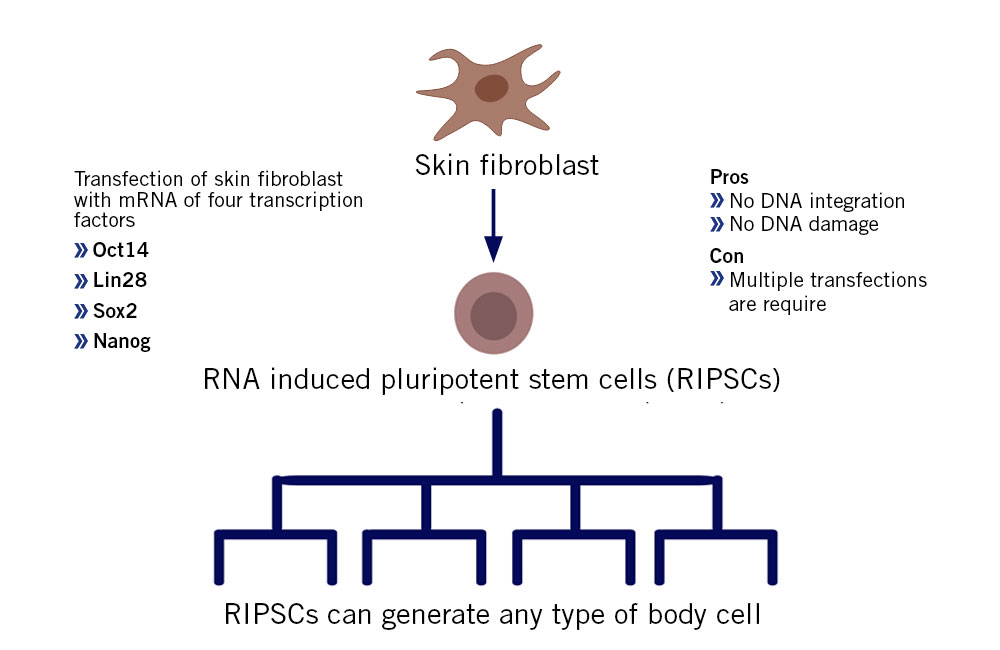
Protein replacement therapies
Fundamentally, mRNA therapeutics can be considered as a transient form of gene therapy that bypasses the complications of “conventional” gene therapy where DNA is inserted in the genome, including insertional mutagenesis and toxicity associated with viral vectors. Currently, researchers are working on introducing mRNA-based protein replacement therapies for treating myocardial infarction (heart attack) (Weissman, 2015).
Genetic engineering
In 2012, teams led by Jennifer Doudna and Emmanuelle Charpentier independently published papers showing how the power of CRISPR can be harnessed to edit the genome. In general, the system is composed of the Cas9 enzyme and a single stranded guide RNA (sgRNA) that are delivered to the cell via a plasmid or viral vector. It has been found that vectors are relatively stable and can persist in the cell. Off-target activity of the CRISPR/Cas system, however, is partly attributed to the sustained persistence of plasmid DNA encoding Cas9 in the cells.
mRNA technology can address this limitation and provide further benefits. sgRNA and Cas9 mRNA can be co-delivered to the cells. This approach has several advantages.
Firstly, mRNA is transiently expressed, which limits the probability for off-target activity.
Secondly, mRNA is directly translated in the cytoplasm. In contrast, plasmid DNA has to cross the nuclear envelope to enable transcription, as plasmid DNA relies on the host’s transcription and translation machinery to express Cas9. The use of mRNA bypasses this step, resulting in a quicker onset of gene editing.
Thirdly, as guide RNA and Cas9 mRNA are both single-stranded, they can be encapsulated in a single nanoparticle, ensuring the delivery of both components into the target cells. Therefore, the mRNA platform can address the limitations of gene editing approaches (Xiong et al., 2018).
Antibody therapy
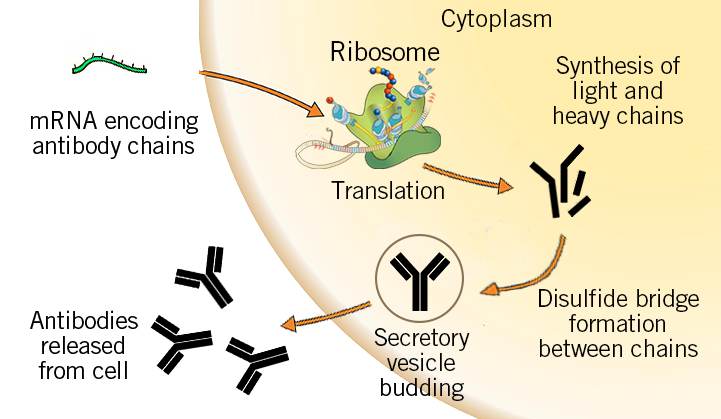
Antibodies are the tailored molecules produced by the immune cells to fight pathogens. Antibodies can be injected into a person’s body as an antidote for a toxin or to clear infections. Antibodies are composed of four chains: two light chains and two heavy chains. These peptide chains are modified and linked together via disulfide bonds.
For antibody production, eukaryotic cells are necessary, as prokaryotic cells cannot glycosylate the peptide chains. This production method, however, is very expensive. To enable cost-effective expression, some researchers have produced single-chain variable antibody fragments in E. coli. These fragments preserved the essential characteristics of antibodies. Unfortunately, their serum half-life is very short.
Antibody mRNA could be a much more suitable and cost-effective alternative. It can be modified easily and transported directly to the patient’s cells. The antibodies are then produced as long as the self-amplifying mRNA is present in the body, providing long-term expression (up to 6 weeks) of specific antibodies (Brito et al., 2014).
1. mRNA encoding antibodies against toxins: Antibodies are administered in people exposed to venom (from snakes and spiders for instance) or toxins (diphtheria, botulinum). Untreated, it takes the body several days or even weeks to produce antibodies. However, in the case of venom or toxin, an immediate neutralising response is paramount. Antibodies that neutralise toxins are produced by using mammalian cell cultures; a very expensive and less accessible system. mRNA technology can facilitate the direct production of antibodies and the administration by intravenous, intradermal, subcutaneous, intramuscular or intranodal route allows for a fast response. mRNA-mediated neutralisation could be a fast acting alternative to the use of antibodies (Schlake et al., 2019).
Since the mRNA needs to be taken up and expressed by the immune cells immediately, much attention has been given to the type of mRNA vaccine used, administration route and delivery format (naked, carrier mediated, cell based). Lindsay and team, working at Georgia Tech and Emory University, tracked the movement of the mRNA vaccine in-vivo. They reported that the first strong signal of mRNA in the lymphoid node was observed after 4 hours as a result of intramuscular administration (Lindsay et al., 2019).
2. mRNA encoding antibodies against infectious diseases: In the past, antibodies were produced by challenging horses with a pathogen and then collecting their serum containing antibodies (polyclonal antibodies). This approach has been utilised to treat respiratory syncytial virus, hepatitis A, hepatitis B, cytomegalovirus, and measles. However, in some patients, equine sera (horse-derived sera) can cause a hypersensitivity reaction. In the search for more efficient antibody production systems, much work was concentrated on producing monoclonal antibodies (mAbs) in the 1990s. mAbs are a brilliant immunisation strategy; they are highly specific against antigens and can be produced in unlimited quantities with no variation among different batches. Also, no animals are involved in the production of mAbs. However, the development and manufacturing of mAbs is costly, and a very high titre of antibodies is required to treat or protect against infectious diseases. A new strategy includes the administration of mAb-encoding mRNA. Significant progress has been made regarding the efficacy of antibody encoding mRNA against HIV and rabies (Pardi et al., 2017; Thran et al., 2017; Schlake et al., 2019).
3. mRNA encoding antibodies against tumours: mAbs can identify and kill cancer cells directly or by activating immune cells to fight cancer. Additionally, some antibodies are specific to both T cells and cancer cells, known as bispecific antibodies. They bring T cells and cancer cells in close proximity and induce the T cells to kill the cancer cells. mRNA-based vaccines for colorectal cancer, prostate cancer, triple negative breast cancer, bladder cancer, pancreatic cancer, esophageal squamous carcinoma, gastric adenocarcinoma, melanoma, and non-small cell lung carcinoma are currently undergoing clinical trials (Miao et al., 2021). However, there are some limitations of mAbs application. They have a short serum half-life, especially bispecific antibodies. In addition, there are manufacturing issues, including reduced stability, and a tendency of mAbs to aggregate. Impurities can also accumulate during the production process. All these limitations can be addressed by direct application of mRNA encoding for mAbs (Stadler et al., 2017; Schlake et al., 2019).
Bringing about post-transcriptional changes with RNA interference
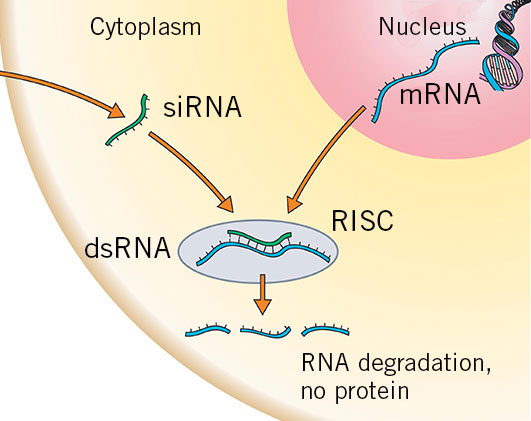
Another class of naturally occurring RNA called small interfering RNA (siRNA) or micro RNA (miRNA) was discovered in 1998 (MK and Kostas, 1998). They are part of the post-transcriptional machinery in the cell. In principal, siRNA and miRNA target specific mRNAs to form double stranded RNA molecules that are quickly degraded (gene silencing). In this way, gene expression in a cell can be controlled at a post-transcriptional level. This approach is used to develop treatments for HIV, cancer and melanoma. Moreover, in 2018, FDA approved mRNA-based therapeutics against hereditary ATTR amyloidosis (DeWeerdt, 2019).
What does RNA hold for us in the future?
The fascinating dream of RNA therapeutics has transitioned to practical reality. In the future, RNA holds the potential to disrupt the field of biopharmaceuticals, vaccine development, cell reprogramming, RNA interference and much more. Read more about where RNA is steering in the future in our the future of mRNA applications article.
Are you working on a mRNA application? Eurofins Genomics can support your research from start to end. Our comprehensive product and service portfolio includes:
Optimised application oligos – Ideal for applications where performance especially matters
By Tamseel Fatima and Dr Andreas Ebertz
Did you like this article on mRNA in biology? Then subscribe to our Newsletter and we will keep you informed about our next blog posts. Subscribe to the Eurofins Genomics Newsletter.







4 Comments