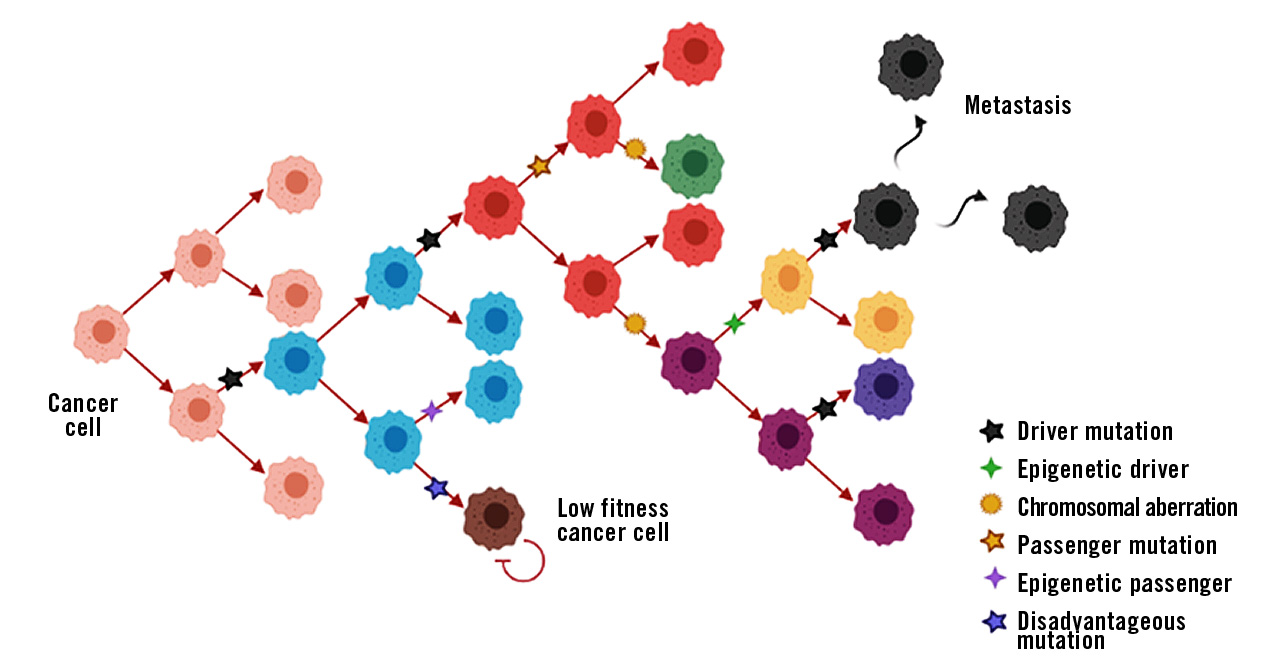
Figure 1: Schematic representation of cancer progression with driver and passenger events. Different cell-colours represent cells with different phenotypes.
Driver mutation: Somatic mutation within a signal transduction cascade that gives the cell a growth advantage and promotes tumour cell proliferation. The largest and most comprehensive database of somatic mutations in cancer is found at COSMIC – Catalogue Of Somatic Mutations In Cancer.
Passenger mutation: Somatic mutation that has no effect on the growth and proliferation of the tumour cell. They are often caused by a mistake in the DNA repair mechanism during mitosis.
What is digital PCR?
The basic principle of dPCR is relatively similar to qPCR and it is also used to quantify DNA sequences. Contrary to qPCR that enables for relative quantification, dPCR enables for absolute quantification.
dPCR shows very high sensitivity which is for instance utilised in diagnosing pathogens and cancer. But how is the high sensitivity of dPCR achieved? The simple answer is partitioning. The amplification reaction of dPCR is similar to that of qPCR and requires the same reagents, such as highly mutation-specific fluorescent probes and primers. The sample that contains a pool of different DNA or RNA fragments is heavily diluted and partitioned into thousands (up to 30,000) of aliquots. These partitions are generated as homogenous droplets on a large 2D array in one single step. Thereby, serial dilutions that are laborious to perform and prone to pipetting errors are avoided.
Subsequently, the nucleic acid fragment of interest is amplified and the resulting fluorescence is detected. The utilisation of several different fluorescence markers also allows for the multiplexing of targets. These advantages make this dPCR method a robust and highly reproducible, as well as effective detection method (Tong et al., 2017; Dhawan and Dangla, 2019).
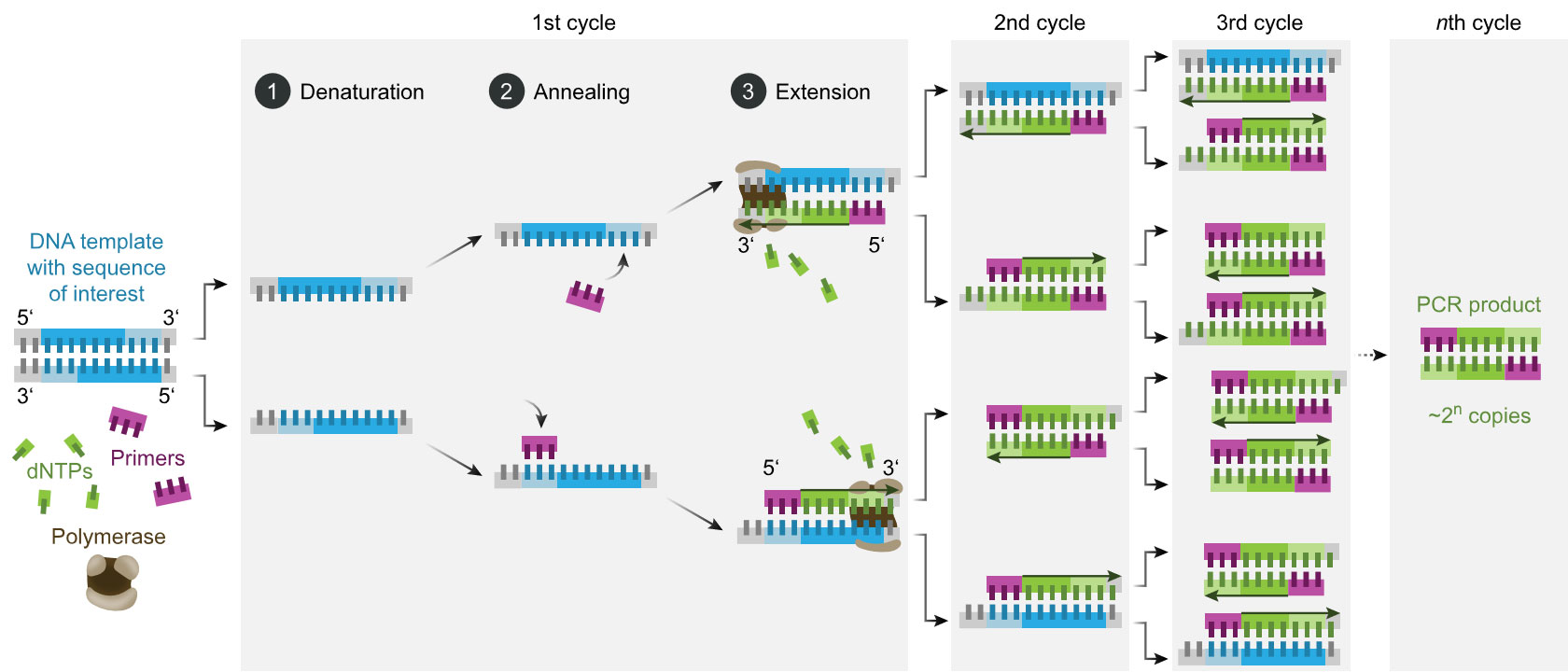
Figure 2a: Schematic of the PCR process, 2b: Image of the Sapphire chip from the naica® system for Crystal Digital PCR™ (image by Stilla Technologies).
In an ideal case, each partition contains either one or no nucleic acid fragment. During the amplification reaction, a fluorescence signal is emitted from partitions that contain the nucleic acid fragment, or no signal if there is no template. The quantification is, therefore, binary (0 and 1), hence the term “digital” PCR. This way, even if only one nucleic acid fragment is present, it is subjected to amplification and detection (Kanagal-Shamanna, 2016).
This shows that the sensitivity of dPCR is staggering! It has been shown that it is possible to detect a mutation rate of 0.001% in a pool of sequences, and a 1.2-fold change in copy number variations (CNVs), making dPCR more sensitive than qPCR.
Furthermore, quantification with dPCR is absolute. As each partition ideally contains only one fragment, and the number of partitions is known, the ratio between positive and negative partitions can be used to estimate the quantity of targeted fragments in the sample (Tong, et al. 2017; Kanagal-Shamanna, 2016).
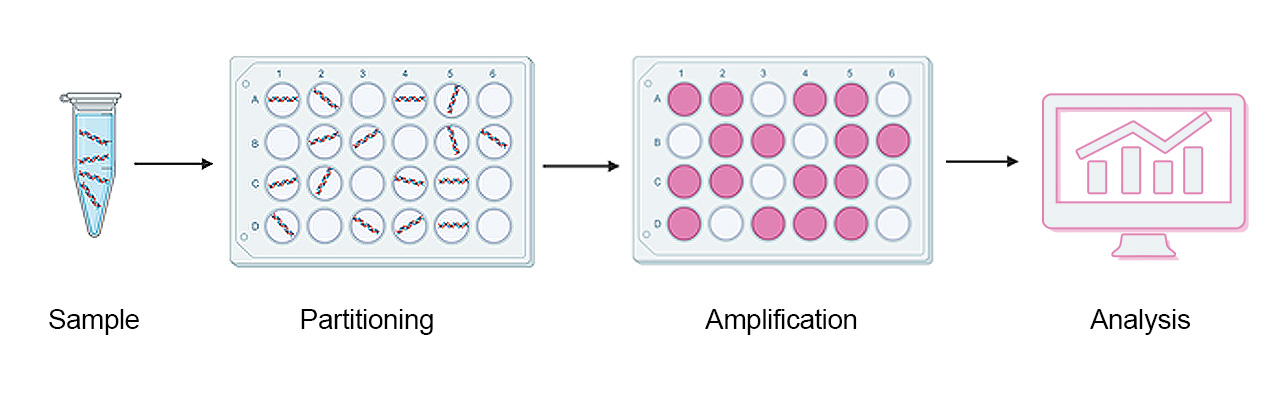
Figure 3: Schematic representation of dPCR workflow.
The perfect couple for cancer diagnostics: dPCR and liquid biopsies
The general rule of thumb in oncology is that the earlier the cancer is diagnosed and treated, the better the outcome. Unfortunately, early-stage cancers are notoriously difficult to diagnose and often go unnoticed. There are many reasons for this, such as lack of symptoms or inadequate techniques to detect tumour markers.
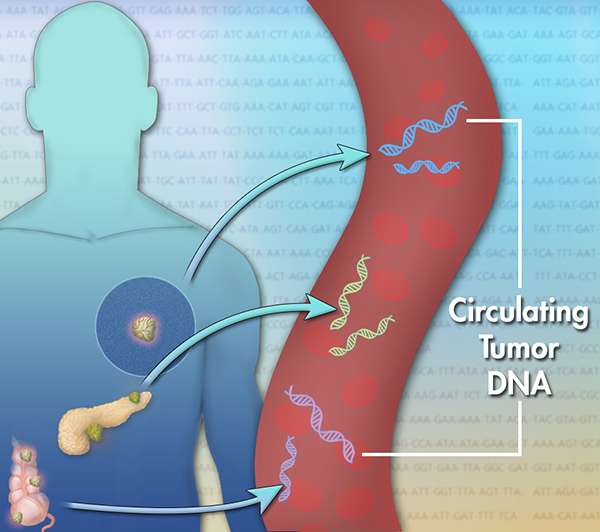
As early stage malignant neoplasms (tumour) secrete different factors into bodily fluids such as blood and cerebrospinal fluid, these so-called circulating tumour markers can be collected and used for diagnostics.
So far, the most widely used circulating tumour markers in diagnostics are protein markers, such as prostate specific antigen (PSA) that is used for detecting and monitoring prostate cancer. However, protein markers can be deceptive and their clinical value is highly debated. The recent advances in biomedicine have put two additional types of marker molecules in the spotlight: circulating tumour DNA (ctDNA) that is part of the cell free nucleic acids – cfDNA and cfRNA – in bodily fluids. These markers can provide a new approach for cancer diagnostics, however, precise and sensitive diagnostic methods are crucial for the detection. cfDNA and cfRNA are usually present only in low amounts in liquid biopsy samples (Postel et al., 2018). For simplicity, the focus of this article is cfDNA and ctDNA.
The genetic changes that occur in a tumour are reflected in the cfDNA that is derived from the tumour (ctDNA). Here, mutated gene sequences can be identified and quantified. The gold standard for diagnostics is qPCR, however, dPCR could be more suitable due to its higher sensitivity. After all, the proportion of ctDNA within the total cfDNA is very small, especially in early-stage cancers. Moreover, pivotal cancer mutations (driver mutations) often occur only on one chromosome, which reduces the amount of ctDNA of interest for diagnostics even further. Consequently, ctDNA could remain undetected in a large pool of wild type cfDNA.
By using dPCR with mutation-specific probes, certain mutations in cfDNA can be detected as early as they start to appear in bodily fluids. Moreover, the high precision and sensitivity of dPCR can be used for monitoring cancer progression and the early detection of a relapse (Tong et al., 2017; De Rubis, et al., 2018).
Liquid biopsy
Liquid biopsy is almost non-invasive, unlike tumour tissue biopsies, and does not require surgical intervention. The advantage of liquid biopsies is that they can be used for regular follow-ups of cancer patients and to analyse the tumour heterogeneity.
Current application of dPCR in cancer diagnostics and progression monitoring
In addition to the usefulness of dPCR to detect certain point mutations, dPCR also comes in handy for detecting CNVs. For example, elevated CNVs in the KRAS gene in patients with colorectal cancer suggests metastasis. KRAS encodes for a GTPase that is involved in signal transduction from the cell surface to the nucleus. Here again, quick identification of these genetic changes could be decisive for therapy outcome (Tong et al., 2017; Vymetalkova et al. 2018).
Lung cancer (bilateral stage IV well-differentiated adenocarcinoma) has been linked to a deletion on exon 19 of the EGFR gene. EGFR encodes for a tyrosine kinase receptor that induces cellular proliferation. The identification of this genomic alteration can aid the selection of a well-suited specific treatment: personalised medicine.
Due to its high precision and sensitivity, dPCR is emerging as a crucial diagnostic tool in oncology. Eurofins Genomics offers attractive dPCR services. Contact our genomics expert and find out how we can support your project.
By Matko Tudor and Dr Andreas Ebertz
Did you like this article on dPCR in oncology? Then subscribe to our Newsletter and we will keep you informed about our next blog posts. Subscribe to the Eurofins Genomics Newsletter.






