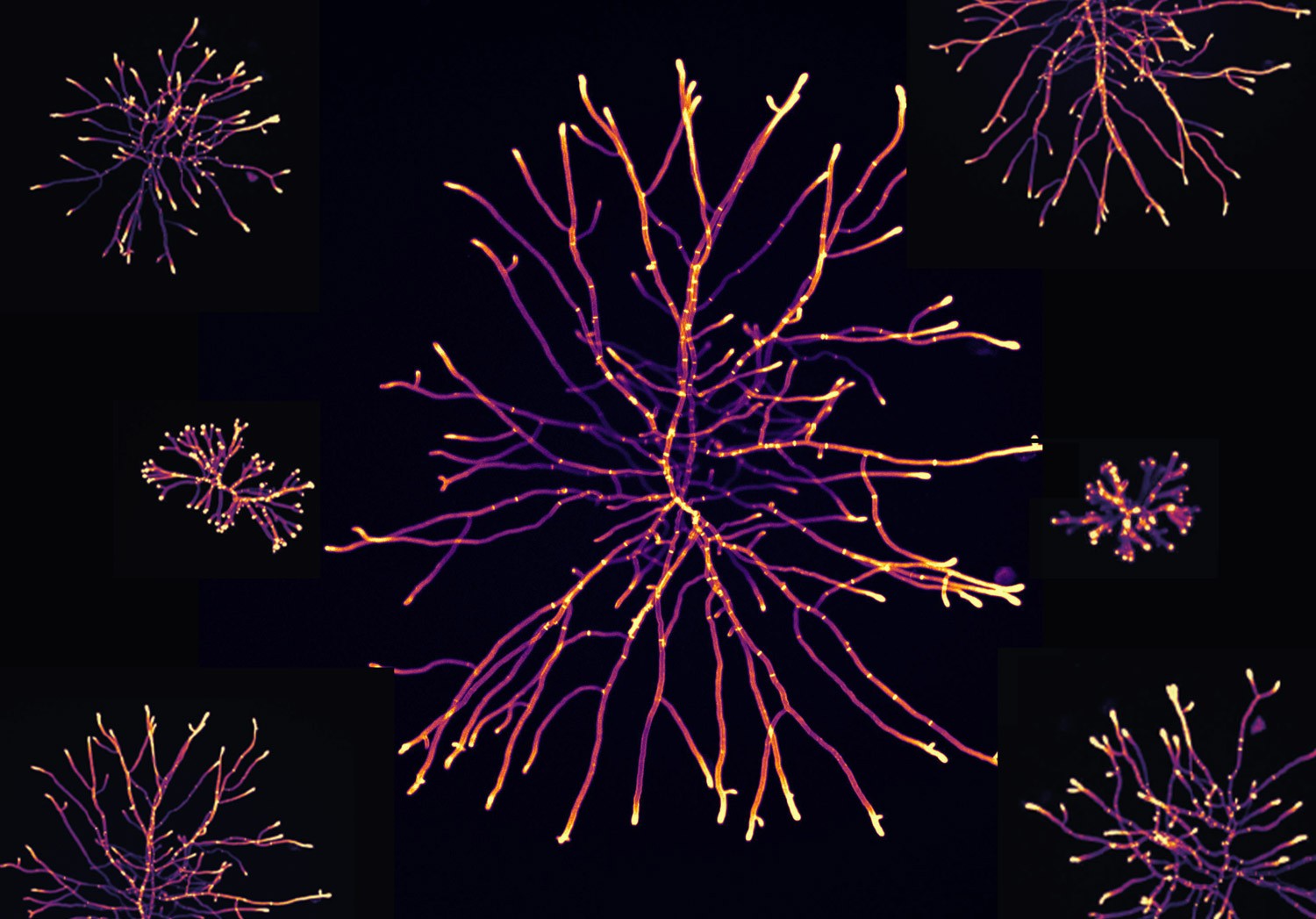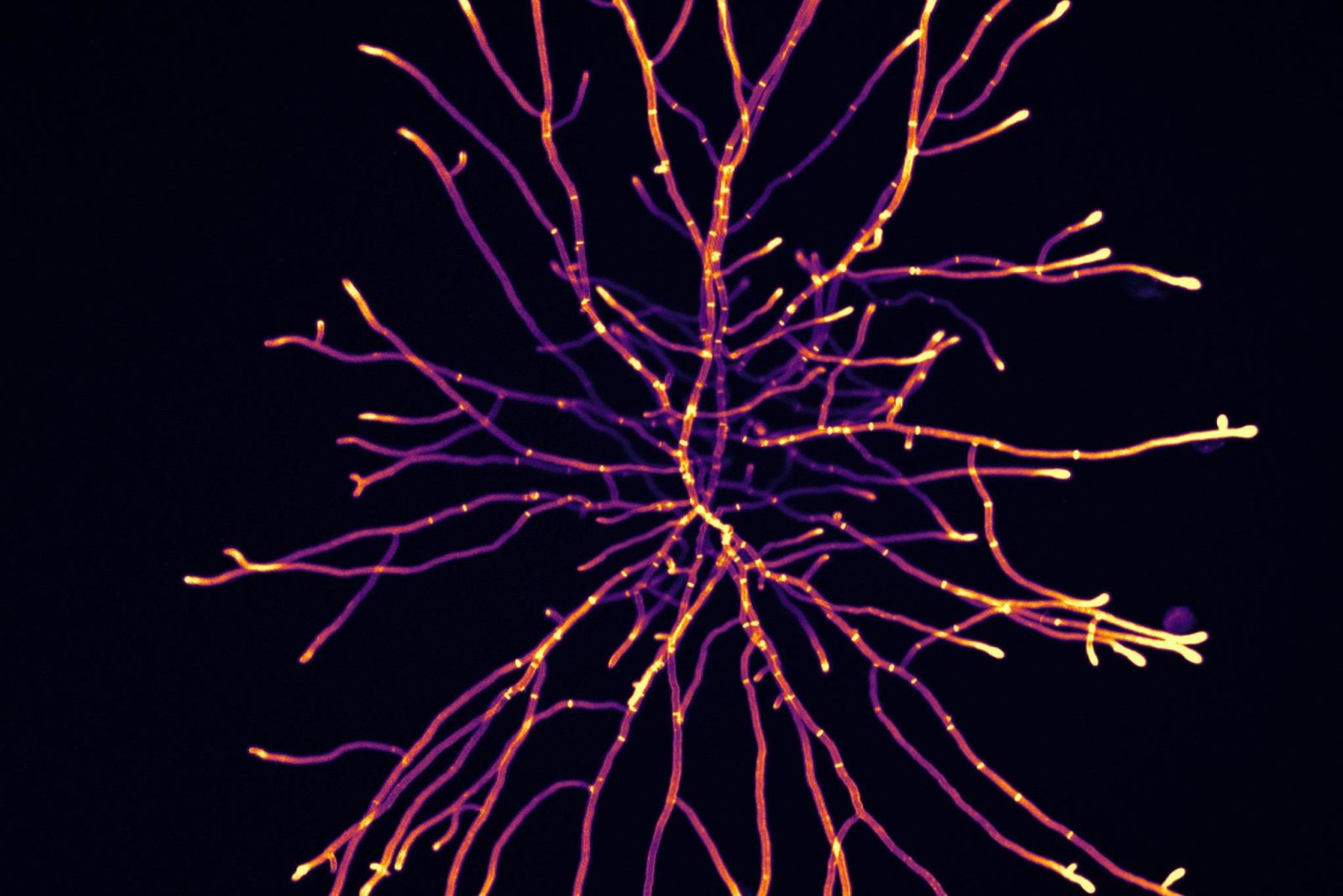
Image 1: The filamentous fungi Aspergillus niger under 100-times magnification. Each thread or filament, called a hypha, and their ability to form branches results in this network of filaments called a mycelium. As a part of the project, we have characterised genetically modified A. niger strains through microscopy. The image was made by staining the cell wall with Calcofluor White and measuring fluorescence. Colour is artificial and scaled to fluorescence intensity.
Our solution to improve A. niger as a cell factory
First, we evaluated which morphological features could be altered to lead to optimal growth in a fermentation tank. A thorough literature research suggested that short and frequently branching filaments would be a possible solution, as most of the protein production takes place in the hyphal tips [1]. The next step was to identify genes that influenced branching regulation in A. niger.
Through extensive literature research, we identified 7 relevant genes in A. niger (gul-1, pkaR, racA, spaA, chsC, arfA and aplD). By utilising CRISPR/Cas9 workflows for A. niger, pioneered at DTU Bioengineering [2][3], we knocked out (K/O) the genes with speed and precision. The genes of interest were first knocked out individually, then subsequently in relevant pairs. We designed sgRNAs (a combination of crRNA and tracrRNA) for the use of the state-of-the-art genetic engineering tool CRISPR/Cas9. Two of our single knockouts and three of our double knockouts were novel in A. niger, and also led to some of our most productive strains – an example is the ∆spaA_∆gul-1 mutant.
Strain characterisation – Who is the best candidate?
After creating 9 strains, we characterised their morphology, growth and productivity by a variety of methods:
- Macromorphology (morphology that can be viewed with the naked eye) was assessed by growing the strains on a variety of solid media to see how the morphology of the strains changes when exposed to different carbon substrates and mineral compositions. For example, some mutants produced relatively few spores on some types of media, which is one way for the cells to channel more resources to protein production.
- Growth rates were examined by using a micro fermentation platform with controlled humidity and temperature. With small reactions, we ran many replicates for each strain across two different types of media (YPD and Minimal media). We also ran larger (1 litre) fermentations for the most promising strain, where we could precisely control stirring, pH and aeration. Among other things, we observed that our productive mutants did not grow noticeably slower than our background strain.
- Protein production was assessed by measuring the specific activity of glucoamylase from the supernatant of the large fermentations. Glucoamylase is a starch-degrading enzyme and was selected for its industrial relevance.
- Micromorphology (morphology at the single-cell level) was assessed from microscope images. For this purpose, we developed an automatic image analysis tool to detect interesting features such as branching frequency. Image 2 is an animation showcasing our image analysis model. We also developed a model to simulate mycelium growth with given morphological parameters, such as the branch formation rate.
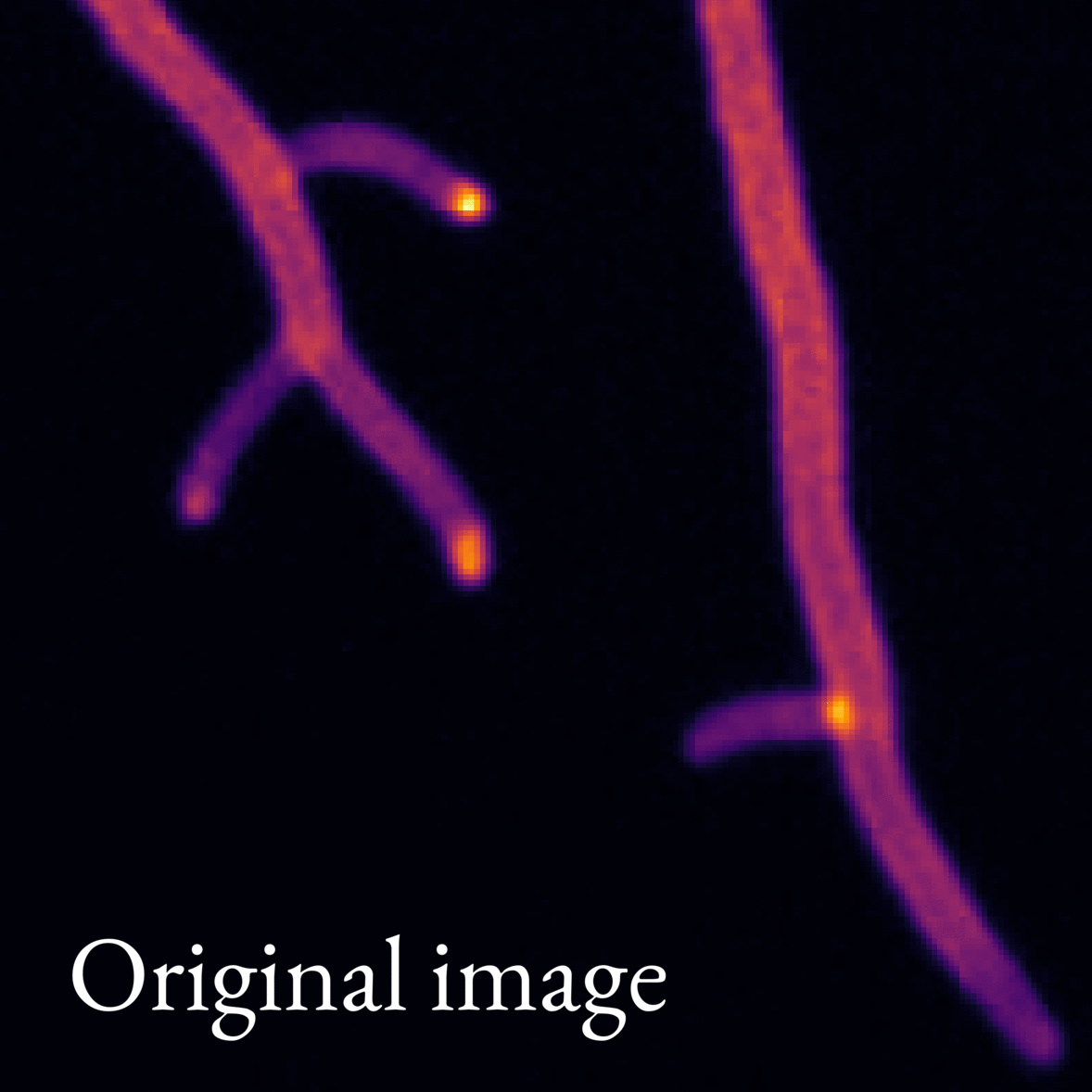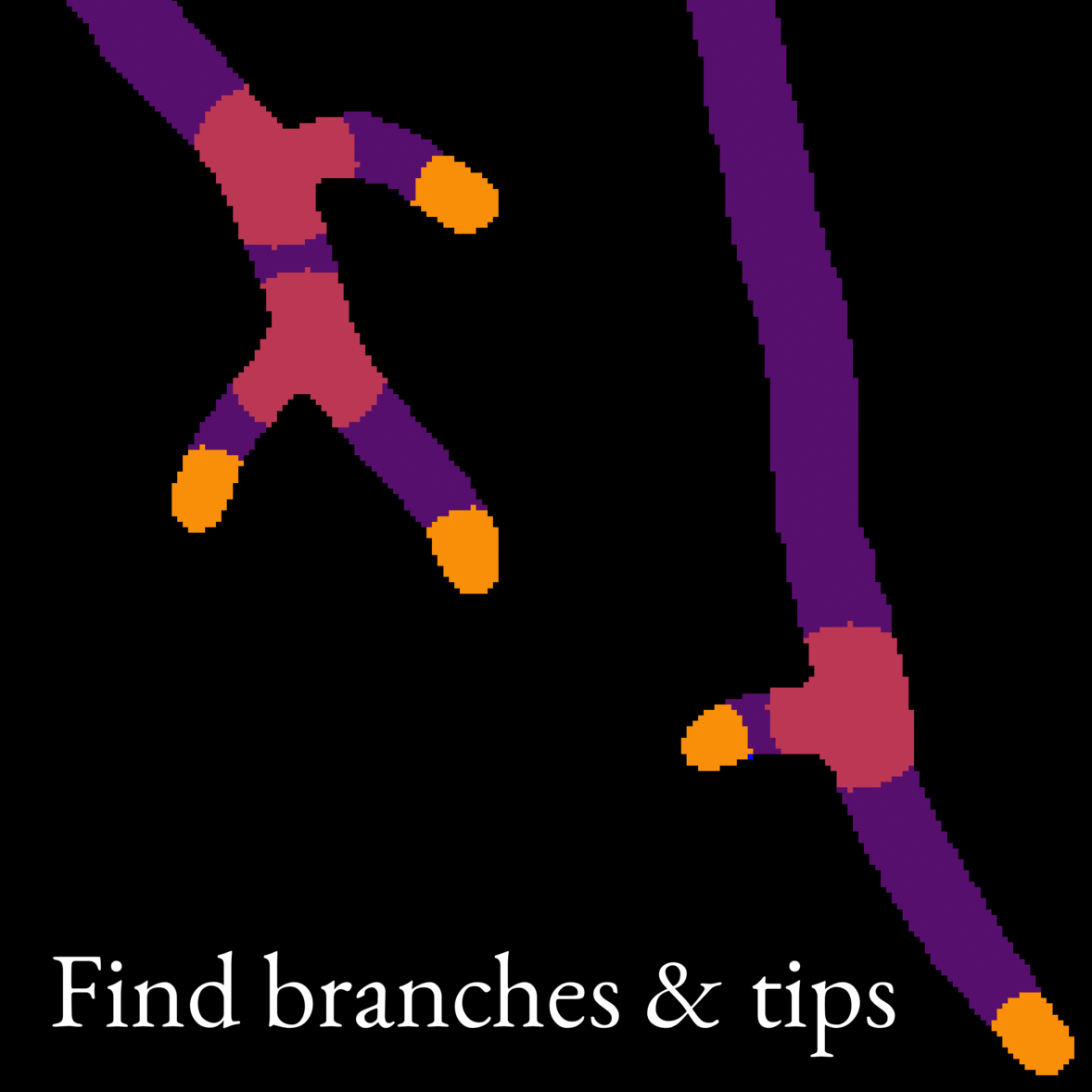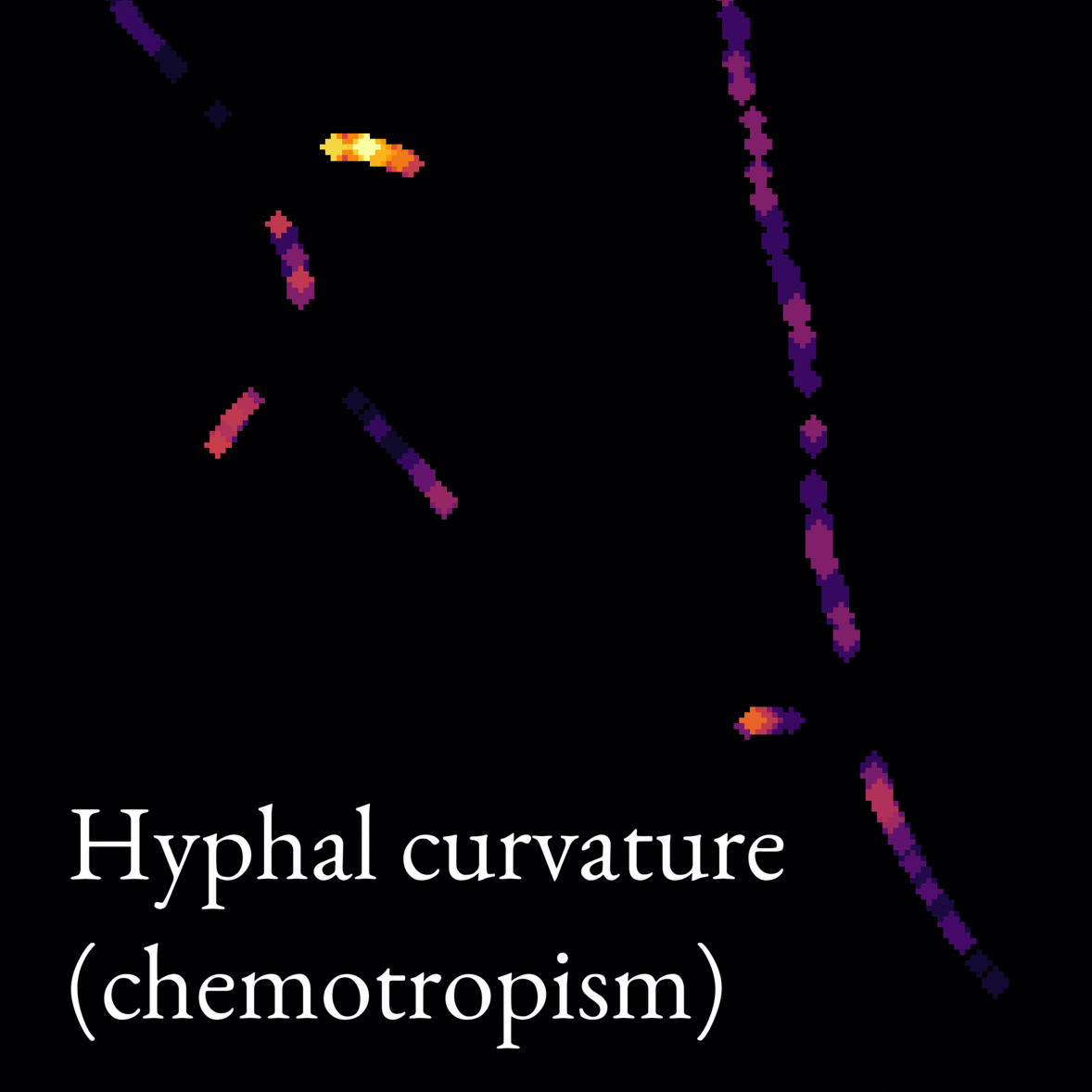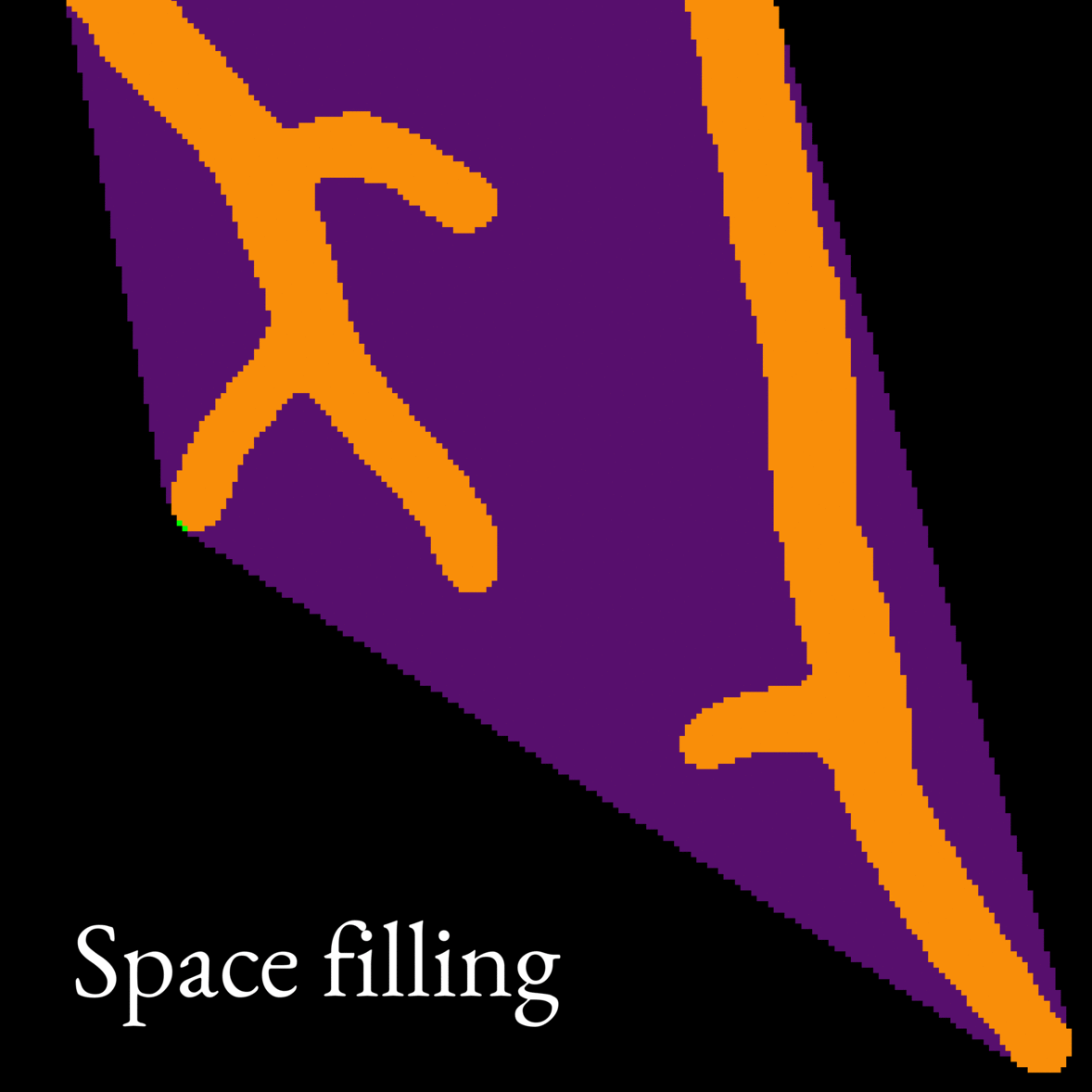
Image 2: Animation showing what features our image analysis model picks out from microscope images of fungi. The model works on whole images of mycelia (like the one in Image 1), but we show only a small section of the image in this animation to illustrate the model better.
Interestingly, our K/O strains showed different growth patterns namely reduced lag phase and increased exponential phases, which could be used to tailor production of future products. For example, some products of the cell are formed mainly during the exponential phase of growth, while others are primarily formed during the stationary phase. This highlights how tuning the duration of growth phases can be important.
The glucoamylase activity was normalised with total protein secretion and quantified with a bicinchoninic acid assay. Fascinatingly, our results indicate that some of our K/O strains have a 2-fold increase in protein secretion and an increase in glucoamylase activity. A chart comparing one of our best strains to our starting strain can be found in image 3. By K/O of these genes, we have shown that it is possible to increase protein production and secretion by tuning morphology-related genes, and, thereby, paved the way for future work with engineering morphology. Among other things, we plan to investigate in greater detail the relationship between productivity and micromorphology.
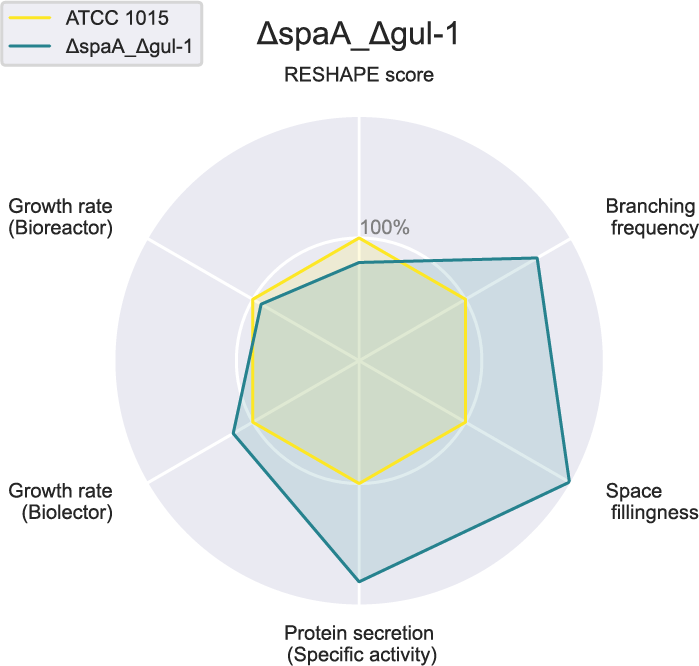
Image 3: Chart showing a comparison between our starting strain and one of our best (most productive and well-growing) mutant strain ∆spaA_∆gul-1. Axes depict the percent increase or decrease compared to the starting strain. Some of our observations, for example the analysis of macromorphology on different solid media, is not included in the chart.
The DTU BioBuilders 2020 Team
We are the DTU BioBuilders 2020 – a student team composed of 13 master students from the Technical University of Denmark, studying Biotechnology and Systems Biology. We participate in the annual international genetically engineered machine (iGEM) competition, which is organized by the non-profit iGEM foundation to promote synthetic biology and collaboration around the world. We have been working on this project in the DTU Bioengineering department since February 2020, and are looking forward to presenting our project at the online iGEM jamboree.

Written and revised by Daniel Bavnhøja, Peter Binga, Clara Drachmannb, Martí M. Gómezb, Cecilia D. V. Graaea, Bira A. Khana, Niels M.a, Lucas Levassora, Margrethe Mærsk-Møllerb, Victoria V. Nissena, Christine Pedersena, Cecilie A. N. Thystrupb, Timian Rindala
aDTU Bioengineering, bDTU Health Tech, Technical University of Denmark

Also check out the iGEM-Team Stuttgart 2020’s project Lac-Man – The Effective Water Filter to Counter Drug Residues in Wastewater.
We are very proud to have supported the next generation of scientist in several European iGEM teams every year. Please have a look at our sponsorship page to learn more about sponsorships opportunities in 2021. Send us a request!