Applications for NGS are much broader than you would think
Next generation sequencing is used to ensure safety and optimisation aspects of large industrial facilities, such as drinking water distribution networks, biogas plants, oil-producing industry and sewage treatment plants.
For drinking water distribution networks, corrosion and iron ochre sedimentation (also termed iron clogging), which is the formation of a thick and gelatinous substance, is partly caused by iron-oxidising bacteria. Iron ochre sedimentation blocks filters and pumps, and reduce the cross-section of pipes, which necessitate more frequent repairs and higher pump performance to deliver the same amount of water. Therefore, timely detection of iron-oxidising bacteria, which is done by metagenome sequencing of the microbial community, could be beneficial.
Also, next generation sequencing can be done for smaller genomes, such as of viruses and bacteria to explore their specific pathways for instance, but also for much larger genomes, such as the human exome to investigate genetic disorders for instance.

New targeted therapeutics are developed based on NGS-results
Next generation sequencing is used to increase our understanding of the genetic factors that are involved in common and rare human diseases. Here, genetic variations are often the cause. A project to map genetic variations called HapMap already collected data of genetic variations in humans from 11 global populations. Genetic factors that are linked to obesity and age-related blindness have already been found. In addition, greater knowledge of genetic variations will also broaden our understanding of how these variations contribute to the immune system, the response to environmental factors, and even the effectiveness of therapeutics (NHGRI, 2010).
Eventually, much more efficient targeted therapeutics that are customised to the individual’s genome and have fewer side effects will be administered.

NGS-based cancer diagnostics tools are developed
Cancer is a cause for significant morbidity and mortality worldwide. Often, cancer detection is the limiting factor when it comes to effective treatment. In recent years, a method for cancer detection that involves liquid biopsies, in fact blood plasma or serum samples, has been developed. The liquid biopsy method is based on circulating tumour DNA (ctDNA) that is released from tumour cells into the blood stream. Instead of using certain genes in ctDNA as markers, the methylation pattern of tumour-derived DNA fragments is used for detection. These alterations occur early in cancer development, they appear more frequently than actual mutations, and occur in specific regions that are called CpG islands (DNA sites with cytosine followed by guanine). The DNA methylation is also chemically and biologically stable, which also makes it ideal for detection purposes.
Investigations showed that next generation sequencing-based analysis of the methylation pattern of specific ovarian tumour-derived DNA fragments in liquid biopsies could be used to detect ovarian cancer up to two years before it is detectable by standard clinical diagnosis tools (Widschwendter et al., 2017a; Widschwendter et al., 2017b; Harper et al., 2016). In epithelial tumours, it has been found that several hundred genes, including tumour suppressor genes, have been silenced by DNA methylation (Wittenberger et al., 2014).
Time to complete next generation sequencing sharply reduced
Sequencing of the first human genome for the Human Genome Project officially took around 13 years and involved researchers from 20 universities and research institutes located in the USA, the UK, France, Germany, Japan, and China. Here, libraries of human DNA were replicated in bacteria – in fact, 20,000 different bacterial artificial chromosomes (BAC) that contained the 3 billion base pairs of the human genome were sequenced (NHGRI, 2010). Sisyphean task anyone?
In comparison, next generation sequencing of a whole genome done in 2019 is achievable within weeks and is done on a single state-of-the-art platform such as the Illumina NovaSeq 6000 with up to 20 billion reads (data output of 2.4 to 3 Terabase of DNA sequence per run with S4 flow cell). Human exome sequencing for instance can be done in 9 business days.

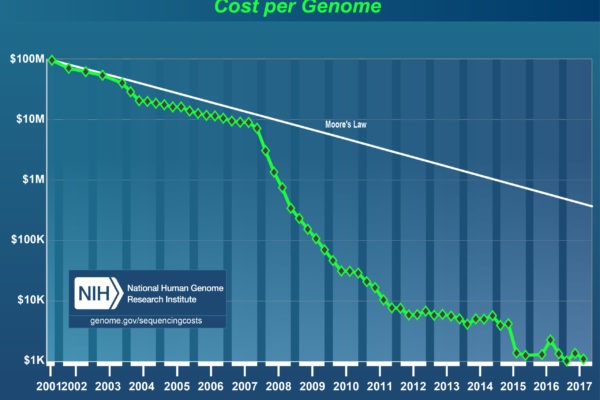
Costs of next generation sequencing
Talking about the Human Genome Project, it cost nearly $3 billion (NHGRI, 2010). In recent years, however, prices for next generation sequencing fell drastically. The cost to determine one million bases of a DNA sequence dropped by a factor of around 100,000 compared to 2001! Similarly, the cost per human-sized genome dropped almost by a factor of 100,000, too (NHGRI, 2018).
Bonus Question
What will be the social and ethical implications of linking DNA variation to individual human characteristics such as intelligence, health status, and temper? Will there be a social restructuring? Will this lead to certain disadvantages for people?
Also stay tuned for more information on the broad applications of next generation sequencing!
By Dr Andreas Ebertz