Home genomics testing
The human body is full of variations that can appear in extreme manifestations, such as unexplained savantism or the unknown pathogenesis of a rare disease, or milder manifestations such as abnormal responses to a therapeutic agent. In any case, the basis of these variations is always founded in genetics. As a result of the great advancements in sequencing methods, precise information about ancestry, predisposition to disease, personality and diet are readily available to the public through home DNA testing kits.
Home genomics testing was first introduced when ‘DNA Solutions’, a DNA testing company, created the ‘home paternity kit’ in 1997 (DNA Solutions, 2021).
The concept behind home genomics testing kits is simple
The home genomics testing kit is delivered conveniently via mail. Once the package with the home genomics testing kit arrives, simple instructions need to be followed to collect the sample. Often, an enclosed swab is used to painlessly scrub cells from the inside of the cheek. The swab is then placed in a tube. Alternatively, saliva is collected in a tube. The collection tubes contain solutions that stabilise the DNA at room temperature. This is crucial, as DNA tends to progressively degrade over time, which makes it difficult to analyse. The sealed package with the sample inside is then returned via mail. Simple!
After the sample arrives in the lab, the DNA is extracted and analysed using microarrays or next-generation sequencing (NGS) technology. The generated data is analysed by a dedicated bioinformatics team and the results are provided online (image 1).
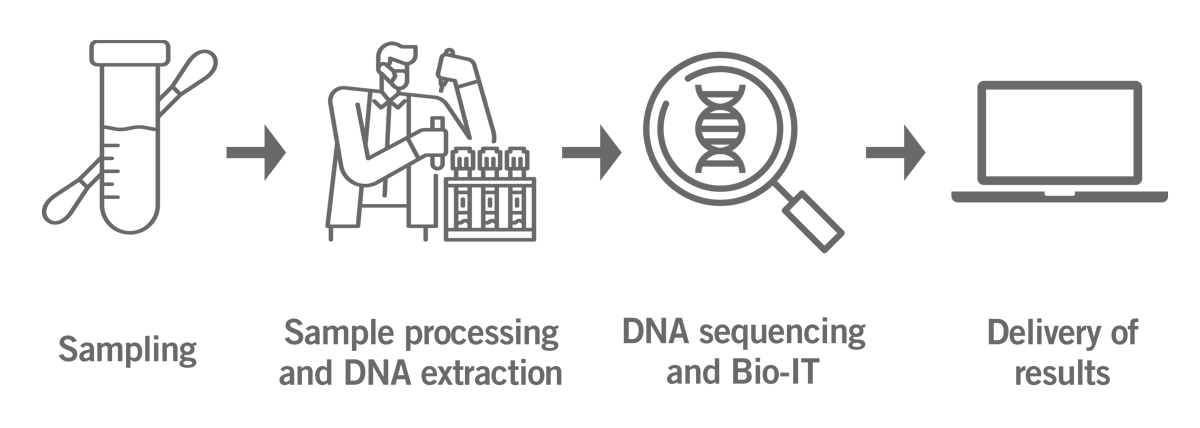
Image 1: Illustration of the home genomics testing process from sampling to delivery of results.
To illustrate how valuable home genomics testing can be, here are some of its applications.
Home genomics testing for personality traits
Many variations in personality traits have been linked to certain genetic polymorphisms. One of the most interesting Big Five personality traits is ‘Openness to experience’ (image 2), defined as the general inclination to indulge in imaginative, artistic and intellectual endeavours. It’s the closest trait that represents intellect and creativity.
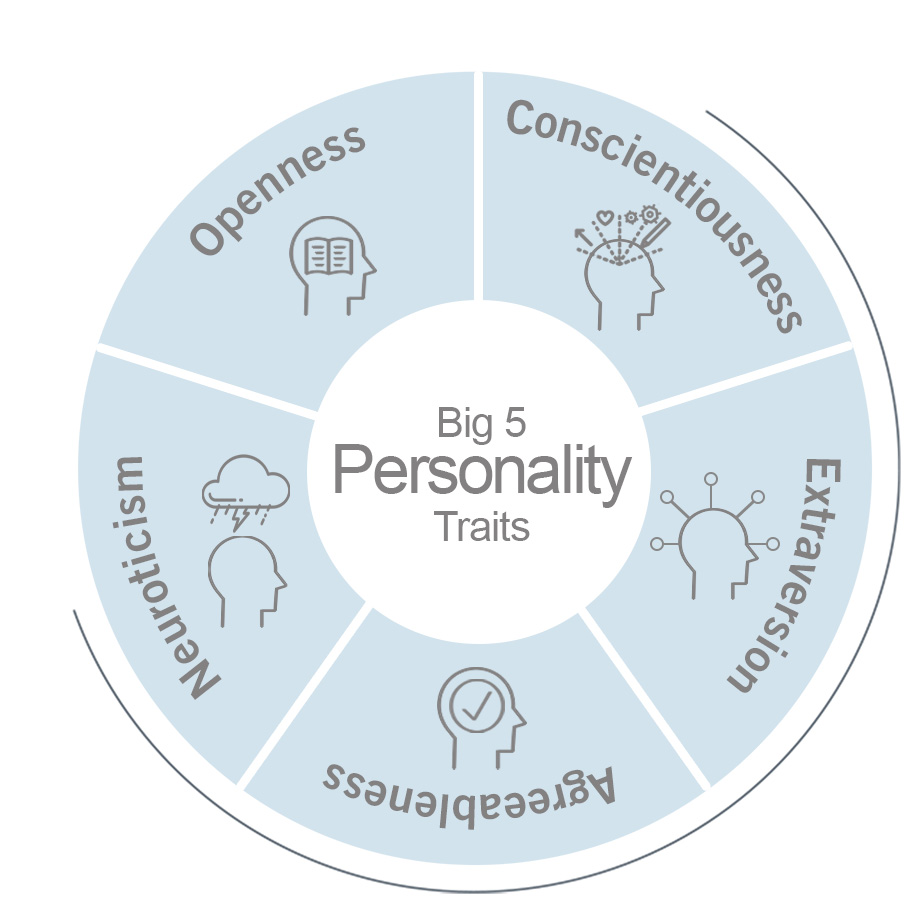
Image 2: Depiction of the big 5 personality traits.
Recent behavioural biology research has uncovered a strong correlation between variations in genes encoding proteins responsible for dopamine metabolism and this personality trait.
It has been postulated that higher levels of dopamine in the prefrontal cortex (PFC) (the part of the brain located in the frontal lobe) (image 3) are associated with higher engagement in cognitive and behavioural exploration and curiosity about both abstract and perceptual information.
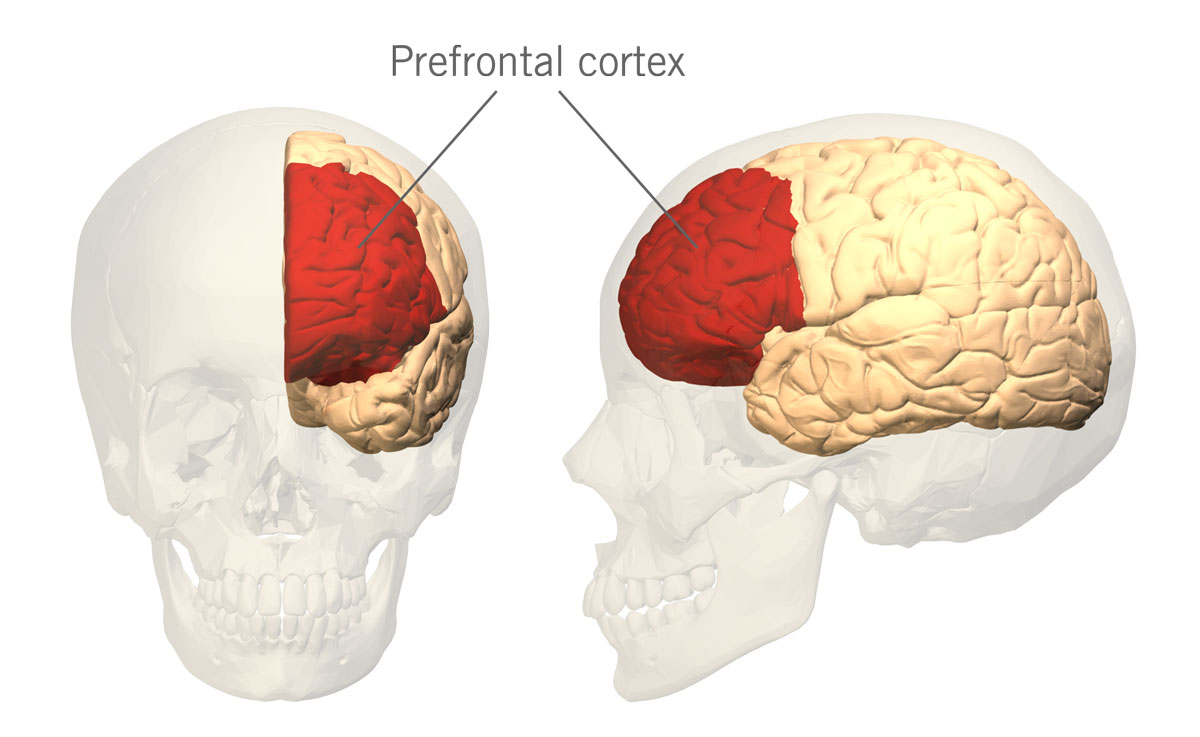
Image 3: Depiction of the prefrontal cortex in the human brain.
Catechol-O-Methyltransferase (COMT) is an enzyme that metabolises dopamine in the PFC and is encoded by the COMT gene. A single nucleotide polymorphism (SNP) called ‘Val158 Met’ has been found in COMT and describes whether a valine or a methionine amino acid has been inserted at the 158th position of the enzyme polypeptide. This rather small change of amino acid in the enzyme, however, has profound effects.
The methionine allele produces a less efficient version of the COMT enzyme, leading to a decreased dopamine breakdown rate and consequently higher dopamine levels in the PFC. This seems to practically translate into higher scores in the personality trait ‘openness to experience’ and thus, could impact the approach to life and behaviour.
Similar to the SNP-based differences in personality traits, physical reactions to different pharmaceutical agents also differ based on the activity of the enzymes that break them down. This activity has been found to be altered by different SNPs.
Home genomics testing for the epigenetic clock – how old are you really?
A person’s chronological age is not necessarily the person’s biological age. People age at different rates, and some people’s biological age appears to be younger or older than their chronological age.
The chronological age represents the number of years of life.
The biological age, in a sense, relates to morbidity and mortality risk, as well as aging according to different clinical biomarkers such as triglyceride levels and telomere length (Belsky et al., 2015).
It is common practice to estimate a person’s biological age using certain clinical biomarkers. However, measuring these biomarkers can be somewhat misleading, as shown by the “Hispanic mortality paradox”, where people of Hispanic ancestry have, in general, higher clinical risk profiles (e.g. risk of diabetes) but live longer than people with European ancestry (Ruiz et al., 2013). According to the epigenetic clock, however, Hispanics seem to age slower.
What is the epigenetic clock? This clock, also known as Horvath’s clock (named after its discoverer Dr. Steve Horvath), is based on the methylation status of DNA (image 4). It has been described as the most accurate molecular measure of age.
The epigenetic clock, and the linked rate of aging, highly depend on the genetic make-up of an individual. It also seems to be hereditary. If a person’s ancestors lived to be 100 years old or more, the probability that this person’s epigenetic clock is “younger” than somebody’s clock whose ancestors did not live to be 100 years old is very likely.
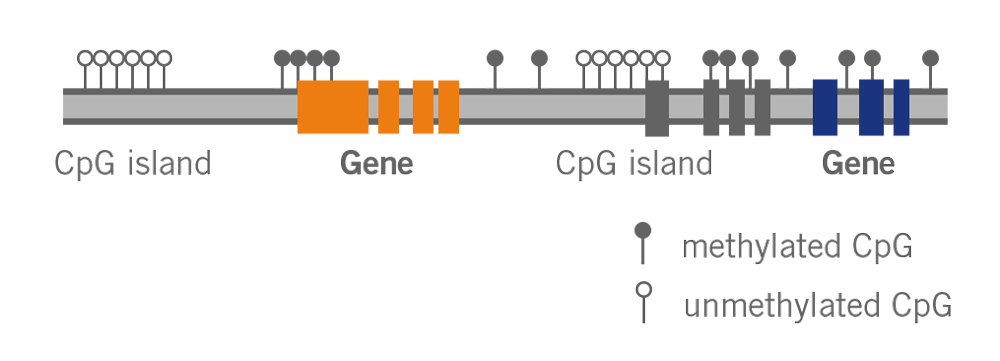
Image 4: Depiction of DNA methylation – methylated or unmethylated Cytosine is found in regions called CpG islands.
What can be estimated or predicted by analysing the DNA methylation of an individual?
With home genomics testing for the epigenetic clock, the biological age compared to the chronological age can be predicted with a very high certainty! If a person’s DNA methylation age is higher than their chronological age, the risk for certain diseases is often higher (positive age acceleration). Conversely, if the DNA methylation age is lower, it implies a lower risk (negative age acceleration).
According to recent investigations, it is possible to statistically predict lifespan and the onset of diseases such coronary heart disease, cognitive decline or cancer based on DNA in blood samples (Lu et al., 2019).
The study also showed the profound statistical effect of diet and lifestyle factors on the biological age (labelled as DNA methylation GrimAge). Eating vegetables and fruits (especially carotenoids), exercising and even education have beneficial effects on aging. Conversely, smoking had the most significant negative effect on the aging of all parameters measured, followed by inflammation (measured by C-reactive protein), a poor waist to hip ratio and diabetes (Lu et al., 2019).
Home genomics testing for drug metabolism
A significant impact of SNPs can also be seen in the genes involved in drug metabolism.
Cytochrome P450 2D6 for instance is an enzyme encoded by the CYP2D6 gene. It is responsible for the metabolism of nearly 25% of drugs on the market, for example some beta-blockers that are used for different health conditions, the most common being hypertension (high blood pressure). In the metabolisms of the beta-blocker Propranolol, CYP2D6 catalyses the oxidation of Propranolol into 4-hydroxypropranolol. SNPs in CYP2D6 account for varying enzyme activity and different phenotypes that are classified using the following descriptions:
- Poor drug metabolisers: Little or no CYP2D6 activity
- Intermediate drug metabolisers: Intermediate CYP2D6 activity between poor and normal metabolisers
- Extensive drug metabolisers: Normal CYP2D6 activity
- Ultrarapid drug metabolisers: Higher CYP2D6 activity that occurs as a result of the expression of multiple copies of CYP2D6 gene (image 5).
Knowing which class of metabolisers a person belongs to and accordingly adjusting the dose is of great importance in order to achieve the optimum therapeutic effect and avoid unwanted side effects.
Image 5: Depiction of metabolisation of drug “X” for poor, intermediate, extensive and ultrarapid drug metabolisers.
Home genomics testing for nutrient and vitamin metabolism
The individual metabolism of vitamins and other nutrients also varies drastically according to the individual’s genetic make-up. For instance, SNPs in genes such as CYP2R1 and CYP24A1 can be utilised to predict whether or not a person is likely to suffer from a deficiency in vitamin D.
Vitamin D needs to go through two hydroxylation reactions to be converted into an active form such as vitamin D3.
The first hydroxylation step is catalysed by the enzyme CYP2R1 (Vitamin D 25-hydroxylase). SNPs in the CYP2R1 gene have been associated with lower 25-hydroxyvitamin D3 levels and consequently lower active vitamin D3 serum levels.
CYP24A1 is the gene encoding a monooxygenase responsible for degrading active vitamin D3. Four SNPs in this gene have been linked to lower concentrations of vitamin D3 (image 6).
Identifying the personal metabolism activity of vitamins and nutrients could support measures for improved physical and mental performance.
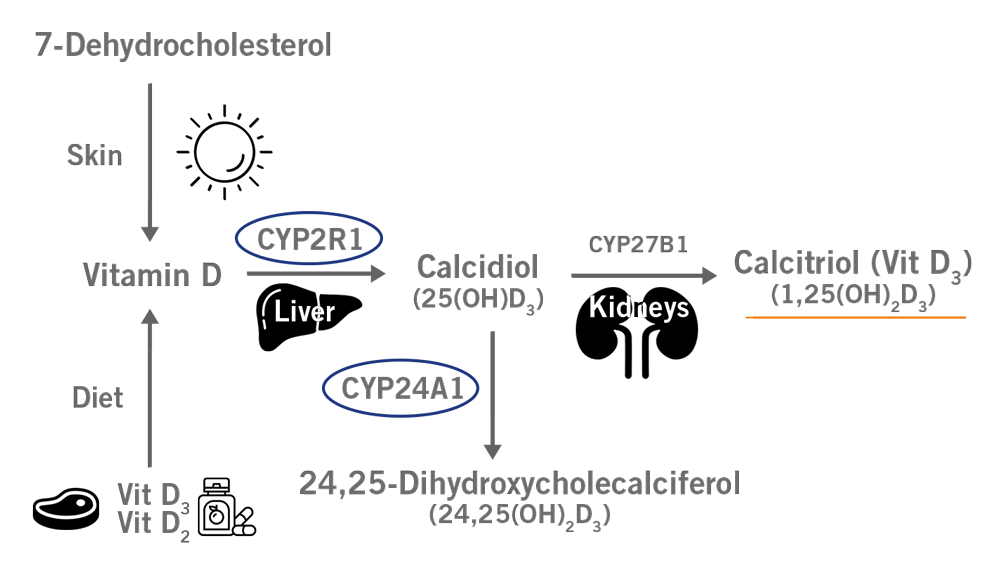
Image 6: Depiction of Vitamin D3 biosynthesis and the involvement of CYP2R1 and CYP24A1.
Home genomics testing of the microbiome
Another useful application for genomics home testing is microbiome analysis. It has been established that the microbiomes of the body, especially the gut microbiome, have profound effects on human health and well-being.
In principal, the microbes in the gut can be classified as commensal/mutualistic, beneficial and pathogenic.
Beneficial microbes are involved in vitamin biosynthesis, dietary fibre breakdown, butyrate production (a highly important compound for mucosal immune functions) and neurotransmitter production (that affect the vagus nerve, a direct connection between the gut and the brain) for example. Commensal or mutualistic microorganisms do not harm the body but seem to be highly involved in the defence against pathogens (Marietta et al., 2018; Khan et al., 2019; Zheng et al., 2020).
This shows the importance of a well-balanced microbial community in the gut. The opposite, a dysbiosis (imbalance of the gut microbiome (image 7), has been linked to the pathogenesis of several illnesses such as inflammatory bowel disease and diabetes.
Even though it seems very difficult to fundamentally change the composition of the gut microbiome, the ingestion of beneficial microbes can have significant effects.
A recent placebo-controlled study showed that a probiotic containing 2 x 1010 CFU Lactobacillus plantarum P8 given daily reduced levels of stress among participants and even enhanced their cognitive abilities and memory compared to the placebo group who received maltodextrin. Lactobacillus plantarum DR7 given daily at a dose of 109 CFU was shown to have the same effect in addition to lowering cortisol levels of participants compared to the placebo group, where plasma cortisol levels did not change (Farahmande et al., 2021).
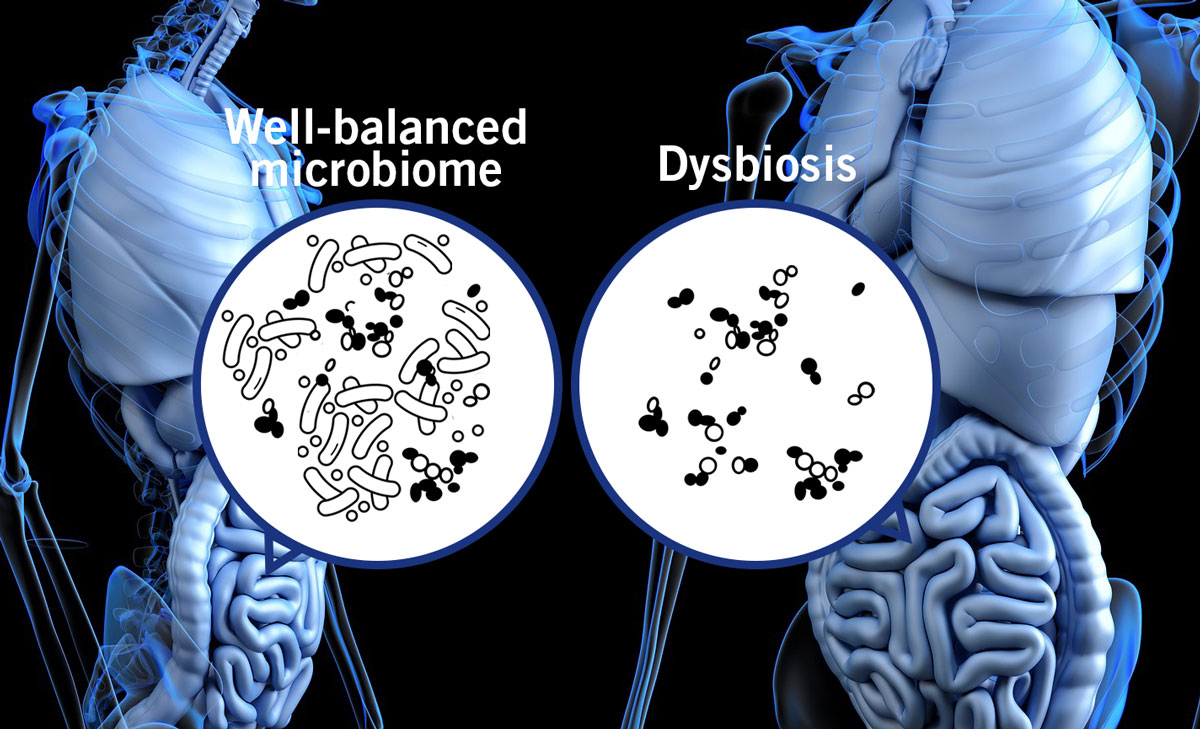
Image 7: Depiction of a well-balanced microbiome and a dysbiosis in the gut.
Gut microbiome home testing kits enable a simple analysis to determine whether a person has a balanced and diverse microbiota that supports optimal body and mind function or not. The analysis results can be used for dietary adjustments to improve the gut flora.
In order to choose the best suited DNA analysis and generate the most informative results, several different technologies are utilised.
Home genomics testing: The technologies behind it
The home genomics testing for personality traits, drug, nutrient and vitamin metabolism is based on microarray technology. Microarrays consist of specific DNA fragments that are fixed to a glass slide for example. These DNA fragments, also called probes, represent the genes of interest, such as the aforementioned gene that encodes for the enzyme CYP2R1, which is vital for Vitamin D3 synthesis. However, the probes vary slightly as they contain SNPs.
The DNA extracted from the person taking the home genomics test is applied to the microarray. The DNA containing the CYP2R1 gene binds to the probes on the microarray. Significantly, the DNA binds specifically to certain probes. If the person has a specific SNP in the CYP2R1 gene, the extracted DNA binds to the corresponding probe on the microarray and not to other probes. Therefore, the person’s CYP2R1 gene variation can be determined and, consequently, it can be estimated how well this person synthesises Vitamin D3. As more than one gene is involved in Vitamin D3 synthesis, several associated genes are analysed by microarray in home genomics testing. This allows the creation of more robust and reliable results.
To estimate chronological, and in relation the biological age, via the epigenetic clock, DNA can be extracted from cells in saliva, urine, blood, or tissue biopsies. For home genomics testing purposes, small amounts of blood are mostly used.
DNA methylation analysis is based on a bisulfite treatment of the DNA. Here, sodium bisulfite converts only non-methylated cytosines into uracils, leaving methylated cytosines unmodified. Downstream processing of the bisulfite-treated DNA includes PCR amplification and the analysis of the resulting amplicons on a microarray that distinguishes between methylated and non-methylated cytosines. The microarray contains all relevant epigenetic clock targets for quantitative analysis of the DNA methylation pattern (image 8).
Cytosine methylation is widespread in humans and is mainly used for analysis. The cytosine in so-called CpG islands is the main point of interest here. Adenine can also be methylated in humans, but is not used for analysis as often.
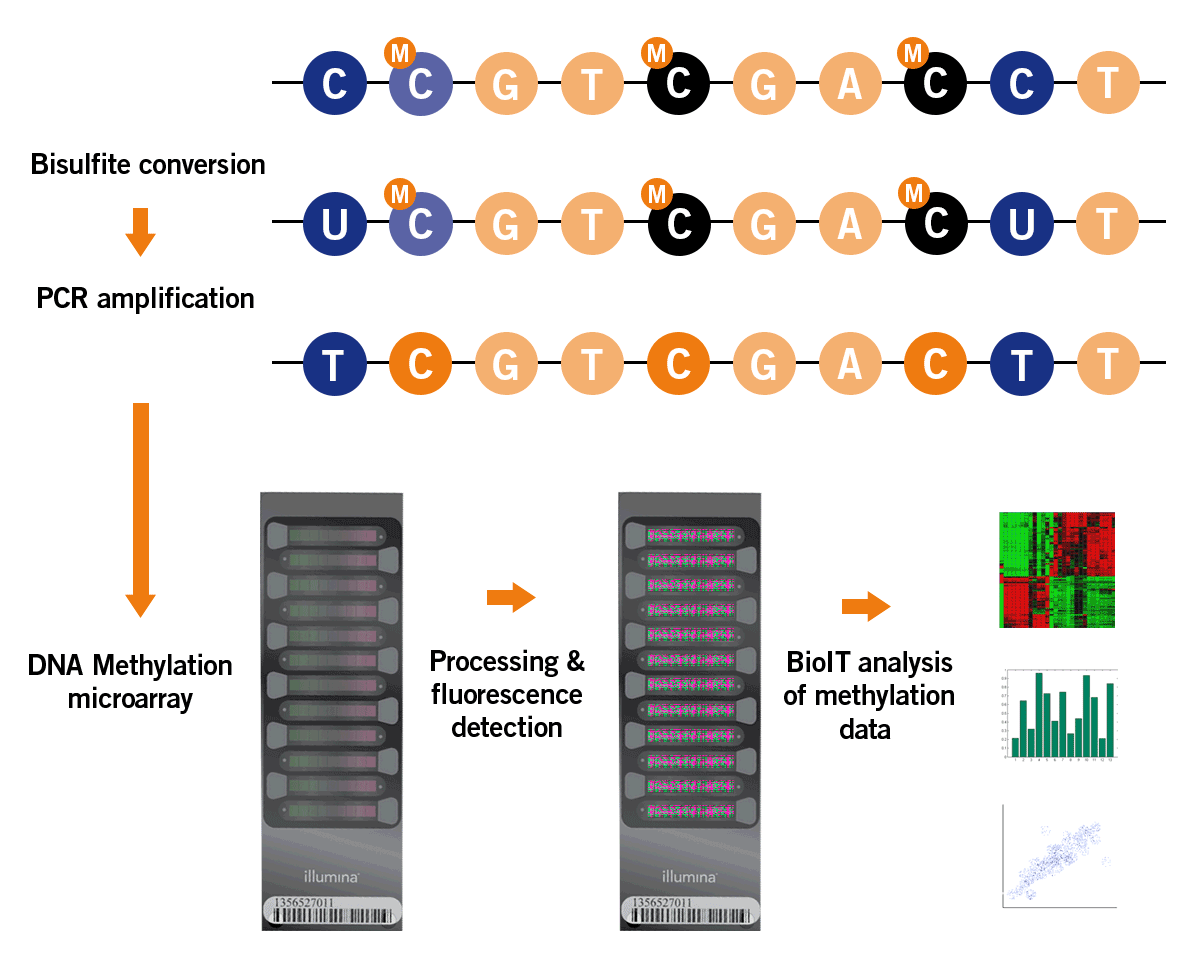
Image 8: Process of DNA methylation analysis by microarray.
The home genomics testing of the gut microbiome is based on sequencing of the 16S rRNA gene of bacteria and the ITS regions of fungi (image 9) or shotgun sequencing (image 10).
Sequencing of the 16S rRNA gene and the ITS region for the identification of bacteria and fungi is the gold standard for microbiome analysis and allows for a comprehensive analysis and identification of the present microbes. For a more details, read our Microbiome Analysis article.
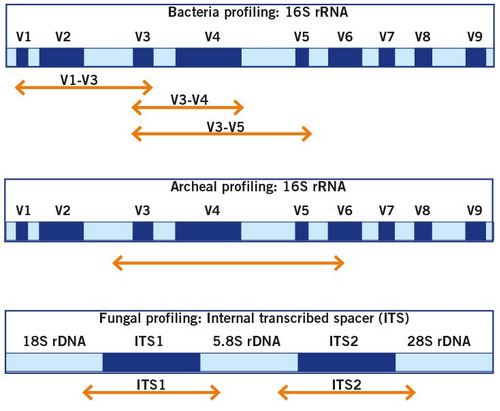
Image 9: Illustration of the genomic regions that are utilised for the identification of the bacterial and fungal community.
Another method of analysing the microbial community is through shotgun sequencing. Here, the DNA from all microbes (mainly bacteria and fungi) in a sample is extracted and randomly fragmented into smaller pieces. These DNA fragments are then sequenced using the Sanger method. The resulting sequences called contigs overlap to a degree. As there are many microbes of every species in the sample and all their DNA is fragmented randomly, the genomes of the different species can be assembled in a following BioIT analysis (assembly) (image 10).
Shotgun sequencing can provide the whole genome sequence of all microbes in the sample, which could allow for a more thorough identification of the present microbes compared to sequencing of the bacterial 16S rRNA gene and the fungal ITS region.
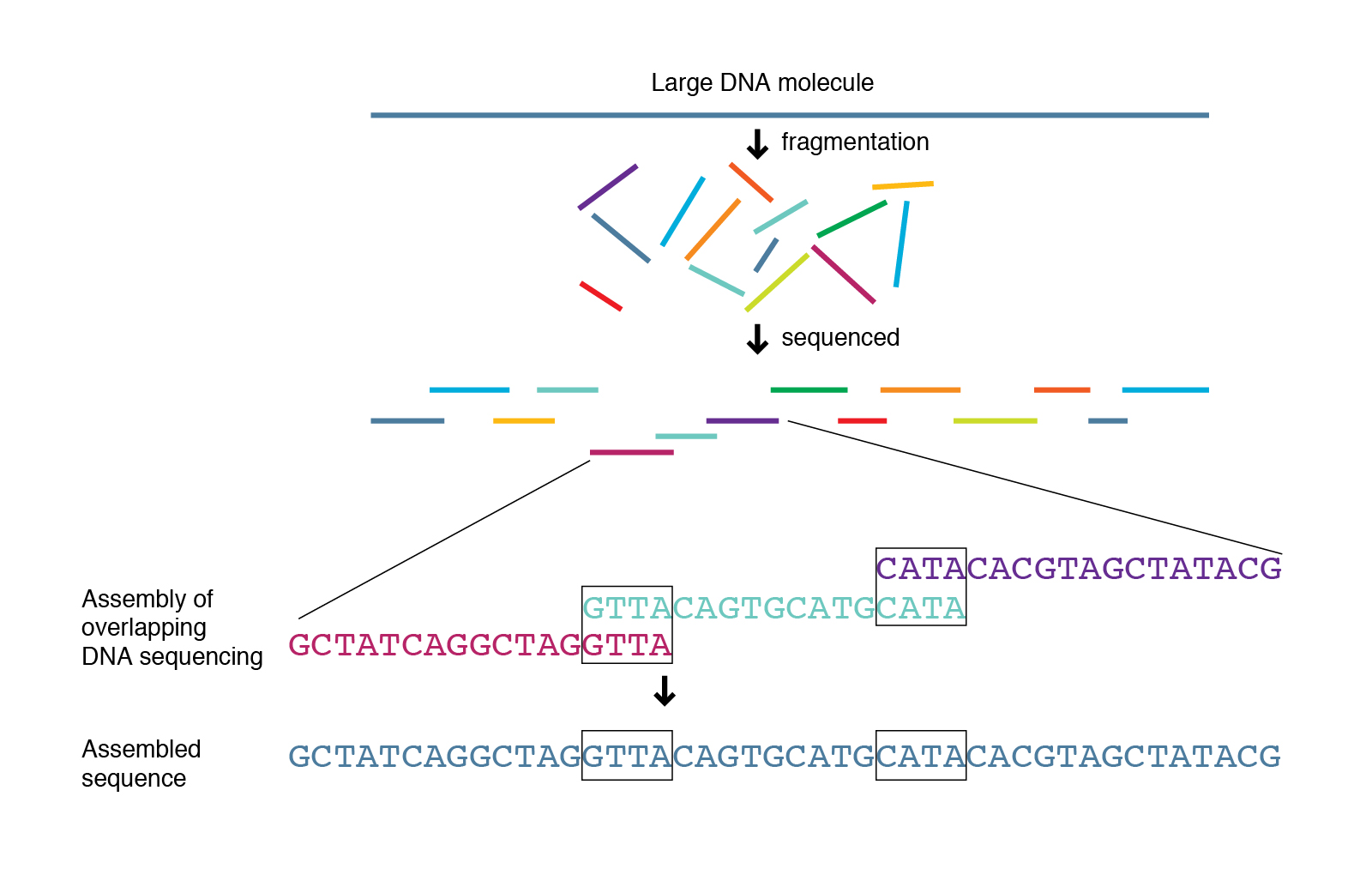
Image 10: Principle of shotgun sequencing.
Home genomics testing for the gut microbiome can also be used to quantify the presence of specific microbial species including viruses. qPCR is the ideal technology for this purpose.
Home genomics testing of microbiomes allows for a wide range of additional applications. Microbiome analysis of the skin, vagina and oral cavity (tongue and gum).
Find out about the Eurofins Genomics services for Home Genomics Testing.
Dr Andreas Ebertz and Abd-ElRahman Samhan
Did you like this article about NGS platforms and capacities? Then subscribe to our Newsletter and we will keep you informed about our next blog posts. Subscribe to the Eurofins Genomics Newsletter.







One Comment