When genes or gene sequences of interest are expressed in a host organism that normally does not possess these genes/gene sequences, it is called heterologous expression.
However, the host organism, also called heterologous system, may not be as efficient in expressing the gene of interest, as the cellular and molecular conditions and signals vary from the one in the donor organism, especially when eukaryotic genes are expressed in prokaryotes.
Codon bias
This is, in parts, based on the redundancy of the DNA code that allows for one amino acid to be encoded by at least two codon triplets. Cysteine, for example, is encoded by the codons UGC and UGU (image 1 A).
However, the frequencies of usage of the codons UGC and UGU differ across species, as seen in image 1B. In the model organism Physcomitrium patens, UGC is the mainly utilised codon for the integration of a cysteine during translation. In contrast, Saccharomyces cerevisiae mainly uses the codon UGU to integrate cysteine. This is called codon bias.
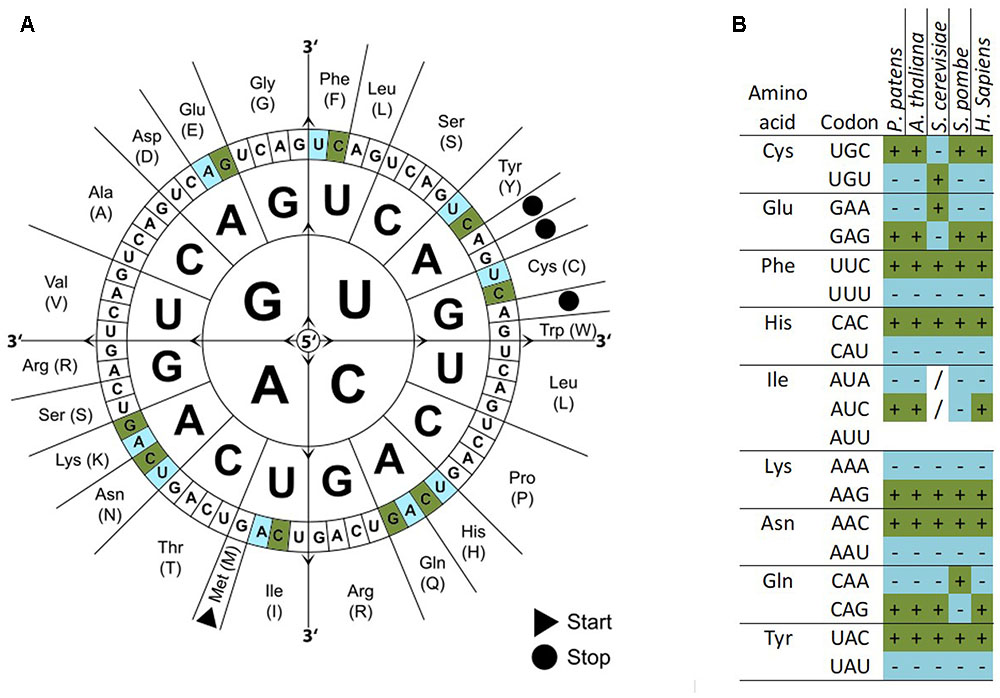
Image 1: A) The genetic code and corresponding amino acid. B) Codon usage bias in model organisms P. patens, A. thaliana, S. cervisiae, S. pombe and H. sapiens. Dark green/marked by (+): codons that are significantly over-represented in highly expressed genes. Light blue/marked by (–): codons that are significantly under-represented in highly expressed genes. (/): codons without bias (Source: https://www.researchgate.net/publication/320729847_Combination_of_the_Endogenous_lhcsr1_Promoter_and_Codon_Usage_Optimization_Boosts_Protein_Expression_in_the_Moss_Physcomitrella_patens, Created by: Manuel Hiss, Lucas Schneider, Christopher Grosche, Melanie Barth, Christina Neu, Aikaterini Symeonidi, Kristian Ullrich, Pierre-François Perroud, Mareike Schallenberg-Rüdinger, Stefan Rensing, Licence: Creative Commons Attribution 4.0 International).
For an optimal heterologous expression, the gene (sequence) of interest needs to be optimised for the codon frequencies of the host organism. Otherwise, codon usage bias could lead to reduced expression rates of the introduced gene due to the limited amount of specific tRNAs, which carry their amino acids to the ribosome.
A codon usage optimisation of a gene sequence should include the usage of codons with high frequencies in the host organism, adjustment of the GC content in the gene sequence and prevention of undesired motifs or secondary structures in the translated protein.
We support the work of scientists who use heterologous expression with our software tool GENEius. It applies a state-of-the-art gene optimisation software and utilises constantly updated databases to provide a comprehensive optimisation solution for synthetic genes.
The GENEius optimisation process includes:
- Optimisation of a gene sequence by employing codons that are naturally used by the heterologous expression system
- Not just the best-suited codons are used, but a combination of codons adjusted to the heterologous expression system
- Prevention of unwanted secondary structures
- Avoidance of direct or inverted repeats
- Removal of hairpins from the final sequence
The GENEius optimisation process excludes bad motifs:
- Avoidance of specific restriction sites
- Omitting of customised “bad motifs” like transcription factor binding sites or splice donor and acceptor sites
The GENEius optimisation process adjusts the GC content:
- Harmonisation of GC content for the entire genes
- Optimisation of GC content in accordance with codon usage table
The GENEius optimisation process insert good motifs:
- Addition of customised “good motifs”
- Facilitation of further subcloning of single protein domains by inserting unique restriction sites
Our GENEius tool outperforms the competition
We tested the optimisation performance of GENEius in comparison to five other software packages from five of our main competitors.
The gene sequence of the jellyfish Aequorea victoria wild-type GFP was optimised using all six software packages for best expression results in E.coli.
After optimisation, the gene sequences were synthesised and expressed in E.coli. The results clearly show higher expression of GFP when the gene sequence was optimised by GENEius compared to optimisations by the tools of our competitiors.
Read the GENEius application note for a detailed analysis of the experiment.
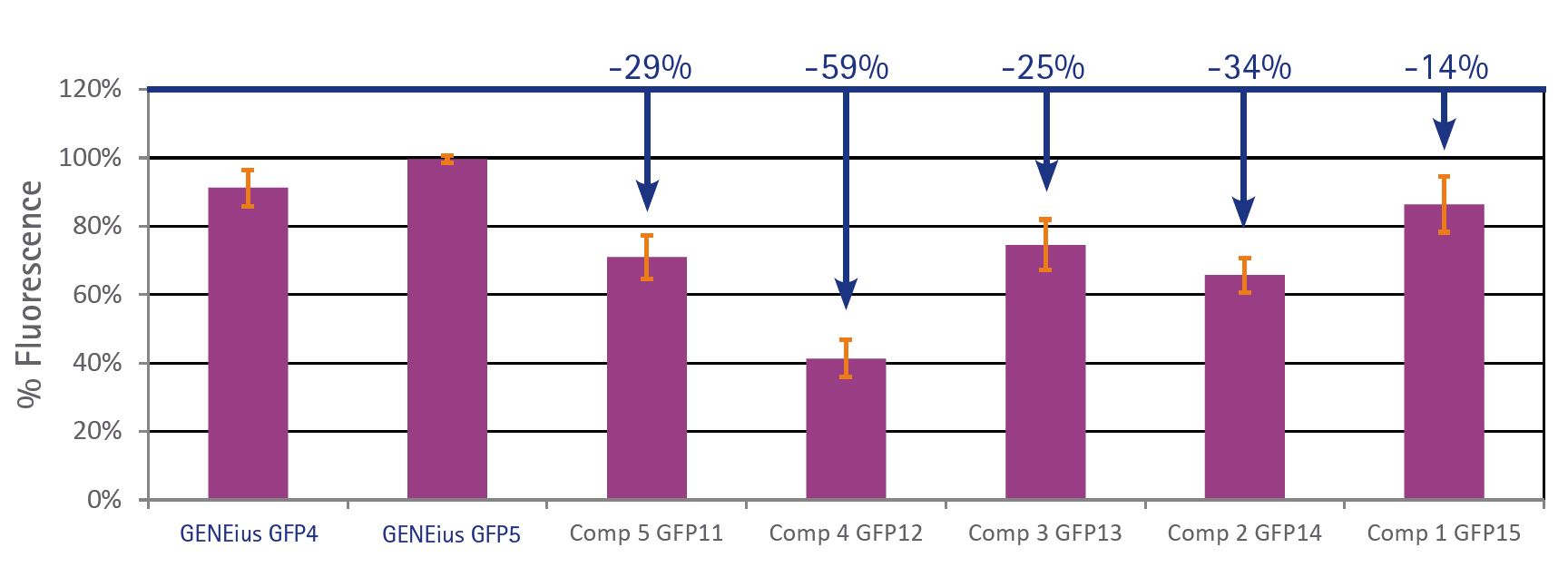
Performance of GENEius
Synthesised genes are often utilised to achieve higher quantities of proteins in heterologous expression systems.
We demonstrated the superior performance of GENEius by adapting a human gene for the
expression in Sf9 cells. The optimisation was performed by comparing the gene sequence to the input codon usage table for Sf9 cells found in the Kazusa Codon Usage Database. For example, the glycine triplet ggg was adjusted by GENEius from a frequency of 0.25 to 0.08 resulting in the optimum frequency for Sf9 cells (0.07).

The expression levels for a GENEius optimised gene in a heterologous system was shown to be strongly increased.

The GENEius tool is easy to handle
- GENEius optimises all protein coding sequences from short DNA fragments of 100 bases to long genes with > 12,000 bases
- GENEius adapts either DNA or amino acid sequences
- Easy selection of species from a dropdown menu for codon usage adaption
- Choose or generate your own “bad motifs” to be excluded, such as cloning sites
- Click “adapt and optimise sequence” to start the sequence optimisation by GENEius
- Our Ecom system will show you the optimised sequence from GENEius
Did you like this article about codon usage optimisation? Then subscribe to our Newsletter and we will keep you informed about our next blog posts. Subscribe to the Eurofins Genomics Newsletter.