There are a lot of myths and misunderstandings about gene synthesis circulating on the internet. Here are 6 myths and facts about gene synthesis you should know.

1. It is impossible to synthesise complete plasmids
This is a myth!
Genes of any length can be synthesised including complete plasmids.
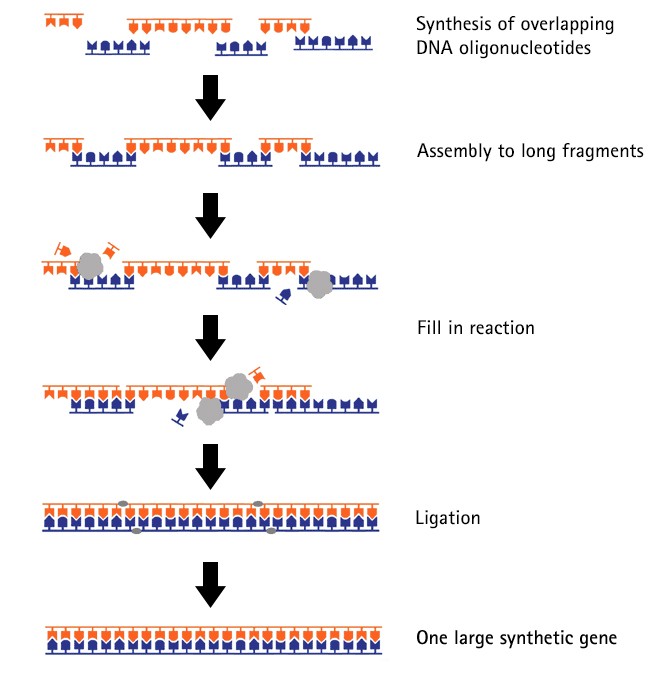
During the gene synthesis process, overlapping DNA oligonucleotides are assembled to long fragments that are then assembled to one large synthetic gene. By adding a circularisation procedure, it is possible to even create complete plasmids of any length.
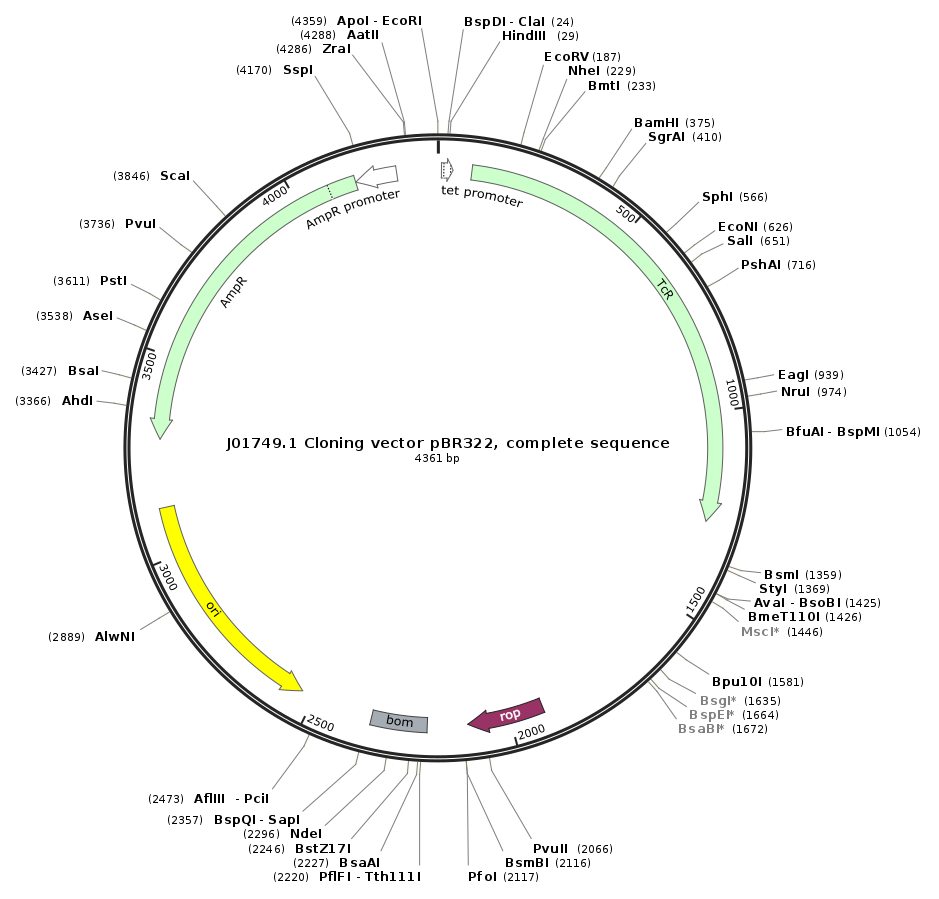
Synthesising a plasmid has several benefits:
- You can freely choose elements such as selection markers, promoter and enhancer, origin of replication, polyadenylation signals, restriction sites, and other.
- You can choose to include a multiple cloning site (MCS) that is tailor-made for your project.
- The nucleotide sequence of the vector is optimised for the host organism in order to regulate the expression levels of genes such as for antibiotic resistance (codon optimisation – see point 3).
- There are no potential licensing issues if you want to use your plasmid commercially.
2. It is impossible to optimise non-coding sequences
This is true!
The redundant nature of the DNA code allows for more than one triplet codon to encode for the same amino acid. Arginine for instance is encoded by the codons CGT, CGC, CGA, CGG, AGA, and AGG. However, the frequency of occurrence of these codons differs across species and is called codon bias. Therefore, it is expedient to optimise a gene for the same frequency of codons as it is common in the organism where the gene will be expressed. Furthermore, codon optimisation contributes to the reduction of high GC content and repeated regions in the gene.
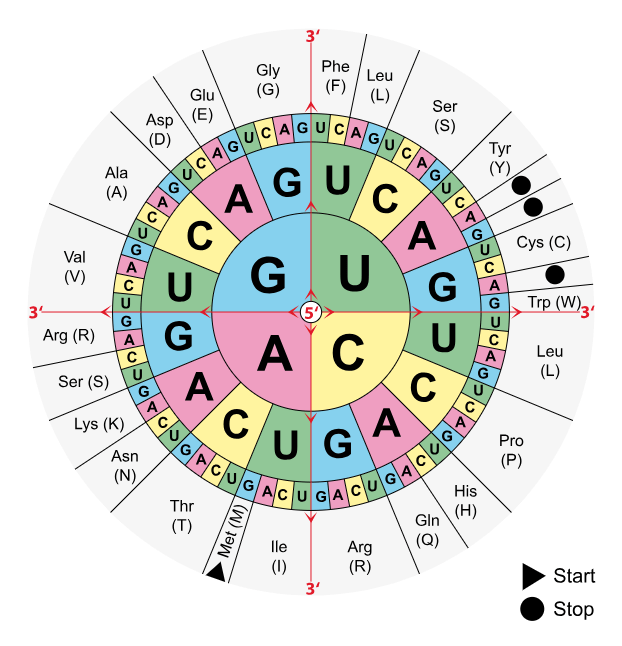
This being said, codon optimisation is only possible when the target gene (or part of it) encodes a protein. Non-coding stretches of DNA do not contain codons that encode amino acids and, therefore, an optimisation cannot be done.
3. Codon optimisation is the only advantage of gene synthesis
This is a myth!
Gene synthesis with codon optimisation is often used for the improvement of the gene of interest in heterologous systems (expression of a gene in a system that normally does not express this gene). Due to codon bias, the expression rates of transfected genes could be lower in the transformed organism compared to the organism of origin, which is based on the limited amount of tRNAs that are available to translate the codons into the respective amino acid sequence. In order to improve gene expression, an adaption of codon usage can be performed with Eurofins Genomics’ gene adaption and optimisation software tool GENEius. It is the best optimisation software at present.
Nevertheless, there are many more advantages for using gene synthesis. For example, when your next-generation sequencing data shows a DNA sequence that you want to investigate further, you can simply order the DNA or RNA sequence. Only the in silico sequence is needed.
Gene synthesis can also be utilised to reliably produce full-length cDNA. A few years ago, labour-, time- and cost-intensive synthesis of cDNA by RNA extraction and RT-PCR was necessary to obtain cDNA. Nowadays, gene synthesis allows for the production and delivery of genes in less than 48 hours.
Another advantage of synthetic genes is avoiding unwanted restriction sites that could occur during subcloning into an expression vector.
4. All gene optimisation tools use only the most suited codons, and a high codon adaption index (CAI) is best
This is a myth!
Simply choosing the most suited codons does not do the trick to yield highest protein expression.
The GENEius tool randomly assembles the DNA sequence and then analyses it in relation to codon usage by comparing it to the input codon usage table that is provided by the Kazusa Codon Usage Database at www.kazusa.or.jp/codon. However, the input codon usage table can also be provided by the customer. Overall, the GENEius tool aims for a high codon adaption index (CAI) AND a harmonisation of the used codons
by integrating the following:
- Frequently used codons from highly expressed genes are more often used for gene synthesis. Very rare codons, however, will be completely avoided.
- GENEius checks for unsuitable motifs, such as restriction enzyme sites, and avoids artificial splice sites, unspecific transcription factor binding sites, etc.
- GENEius avoids the integration of direct and inverted repeats, as they complicate the gene synthesis process, decrease DNA stability, and reduce transcription and translation efficiency as shown in E.coli.
- GENEius ensures equal distribution of the GC content in order to avoid GC-rich sequences within the gene.
All these parameters are reflected in the “optimisation score” shown in the GENEius tool.
5. Even if restriction enzyme sites from the multiple cloning site (MSC) of your vector are missing in your insert, the insert can still be subcloned into the vector
This is true!
You only need to cut your vector with one or two suitable restriction enzymes that target the MCS. Your gene insert can then be PCR amplified with primers to generate regions at the ends of the PCR product that are homologous to the vector. These homologous regions are then used for subcloning via sequence and ligation independent cloning (SLIC) into your vector. Any restriction sites within your insert do not matter at all because the PCR product will not be cut during subcloning. SLIC is as fast and efficient as traditional subcloning via restriction enzyme sites. The Eurofins Genomics’ molecular biology experts will ensure that all your requirements are met and that necessary elements such as a stop codon or a fusion tag at the end of your gene are included.
6. If the standard vector contains sites for my restriction enzymes, there will be issues with downstream cloning
This is a myth!
The Eurofins Genomics’ standard vectors have been designed to contain only few restriction enzyme sites in the MCS. Even if a site for your restriction enzyme is present, this will not cause a problem. After restriction enzyme digestion, the resulting fragment will be so small that it will not be visible on an agarose gel. All restriction sites in the MCS will be at least 4bp (in most cases >10bp) away from your gene so the efficiency of the restriction enzyme digestion will also not be affected.
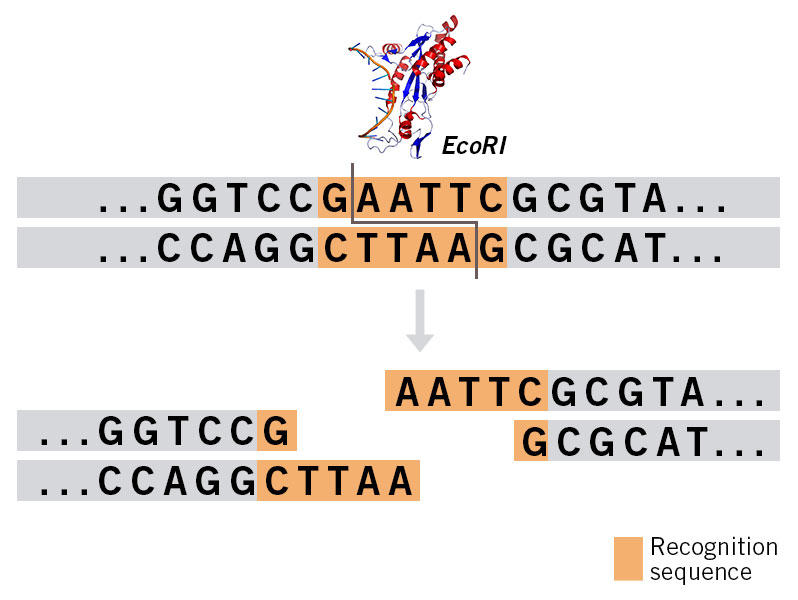
Even in the unlikely event that one of your chosen restriction enzyme sites is present in the vector’s backbone and after the digestions you find a band of the same or similar length as your gene, here are options to get around this problem:
- Use a third restriction enzyme for the digestion to make an additionally cut in the vector backbone and thereby reduce the size of the unwanted band. In most cases, the required band can then be extracted from the agarose gel.
- In case of a lack of suitable restriction enzyme sites, the gene can be amplified with PCR primers. Subsequently, the PCR product is cut out with restriction enzymes and used for subcloning. Here, it is important to have 2-3 nucleotides in front of the 5´ ends of the PCR primers as overhangs so the efficiency of the digest is not reduced.
Bonus: A tip for saving time and money
During gene design, include a promoter sequence upstream of your gene (and a terminator sequence at the 3’ end), and then use the Eurofins Genomics’ pEX vector. The pEX vector is based on pUC18 vector and per se does not contain a promoter or terminator for expression of a subcloned gene. However, due to the inclusion of a promoter sequence upstream of your gene, the pEX vector can be used as an expression vector for protein expression in vivo or, and, after linearisation, as template for in vitro transcription.
Note: The pEX vectors are standard cloning vectors for E.coli.
By Uwe Köhler and Dr Andreas Ebertz





