
Fig 1: Difference between human primary cell culture and cell line (Oyeleye, O. et al., 2016)
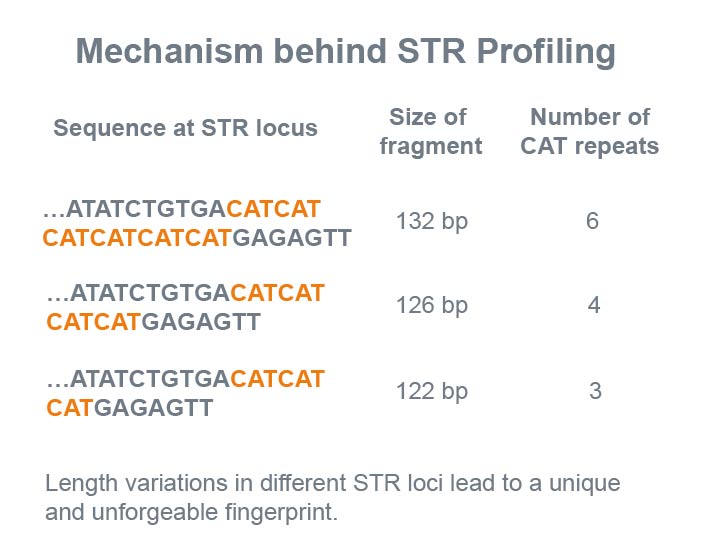
Fig 2: Illustration of the principle behind using STR profiling for DNA fingerprinting
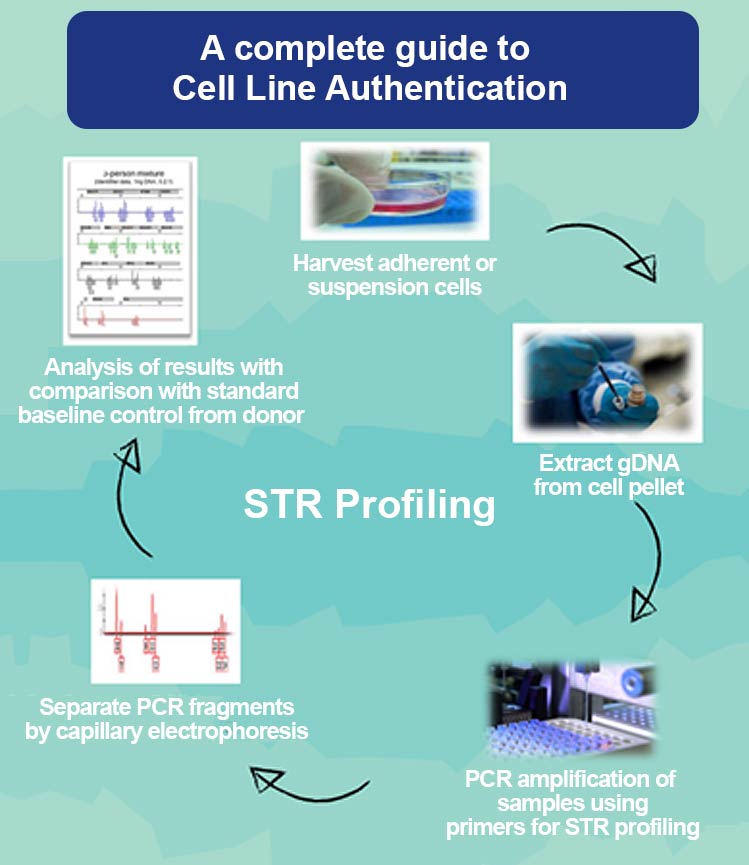
Fig 3: An illustration of the major steps involved in STR profiling.
By Tamseel Fatima and Dr Andreas Ebertz





