We have a dream
We, the 2019 TU Delft iGEM team, want to design a toolkit that allows us to express genetic constructs in different microorganisms, and thus accessing the diversity of bacteria beyond E. coli and S. cerevisiae. In order to carry out our project called Sci-Phi 29, we will rely on genetic systems with orthogonal transcription and replication. Those systems can carry out transcription and replication processes independently of the host’s specific machinery and regulating elements.
This hybrid system that we develop will be able to replicate and transcribe a gene of interest independent of the host’s transcription machinery. For it to work in different bacterial strains, we will also develop a software tool to find consensus sequences for codon usage. We aim to achieve similar levels of expression amongst different bacterial species with the same genetic construct.
This is the toolkit to make our dream come true
To achieve orthogonal transcription, we will develop an expression system that consists of T7 promoters with different expression levels (Figure 1). These will behave similarly in different bacterial species due to a TALE protein regulating system that modulates the activity of the T7 promoters (Segall-Shapiro et al., 2018).
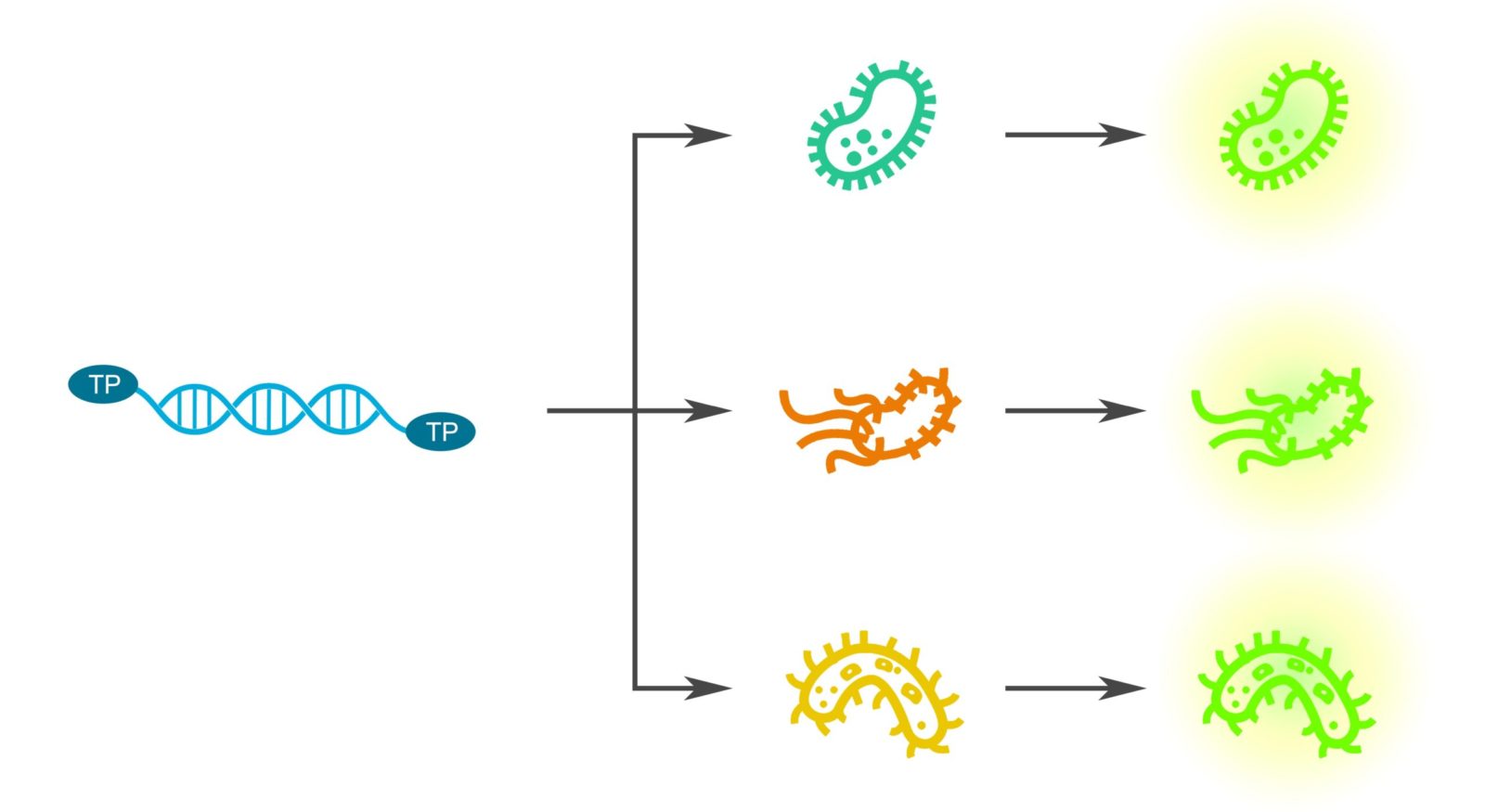
Figure 1: The goal of our toolkit is to create a universal gene expression platform that would enable genetic engineering of any prokaryote.
Additionally, orthogonal replication will be achieved by mimicking the replication system of the bacteriophage phi29. This bacteriophage has a linear genome capped by two terminal proteins. It is duplicated by a single-subunit polymerase that uses the terminal proteins as replication starting points (Figure 2).
We will test our construct in E. coli as a proof of concept and also in the less known bacterial species Vibrio natriegens and Pseudomonas putida. Both of the latter species are very interesting from a metabolic point of view. V. natriegens is able to grow at extremely high rates, while P. putida has a very high tolerance to stressful environments, and can degrade aromatic compounds and other hydrocarbons that are waste products from polluting industries such as the chemical industry.
To wrap up, the Sci-Phi 29 project aims to bring many other bacterial species into the field of synthetic biology. An increase in the variety of chassis and ready-to-use bacterial strains will allow scientists to genetically adapt more hosts, some of them with potentially relevant applications in bioremediation, healthcare, industry, or even energy production in the future.
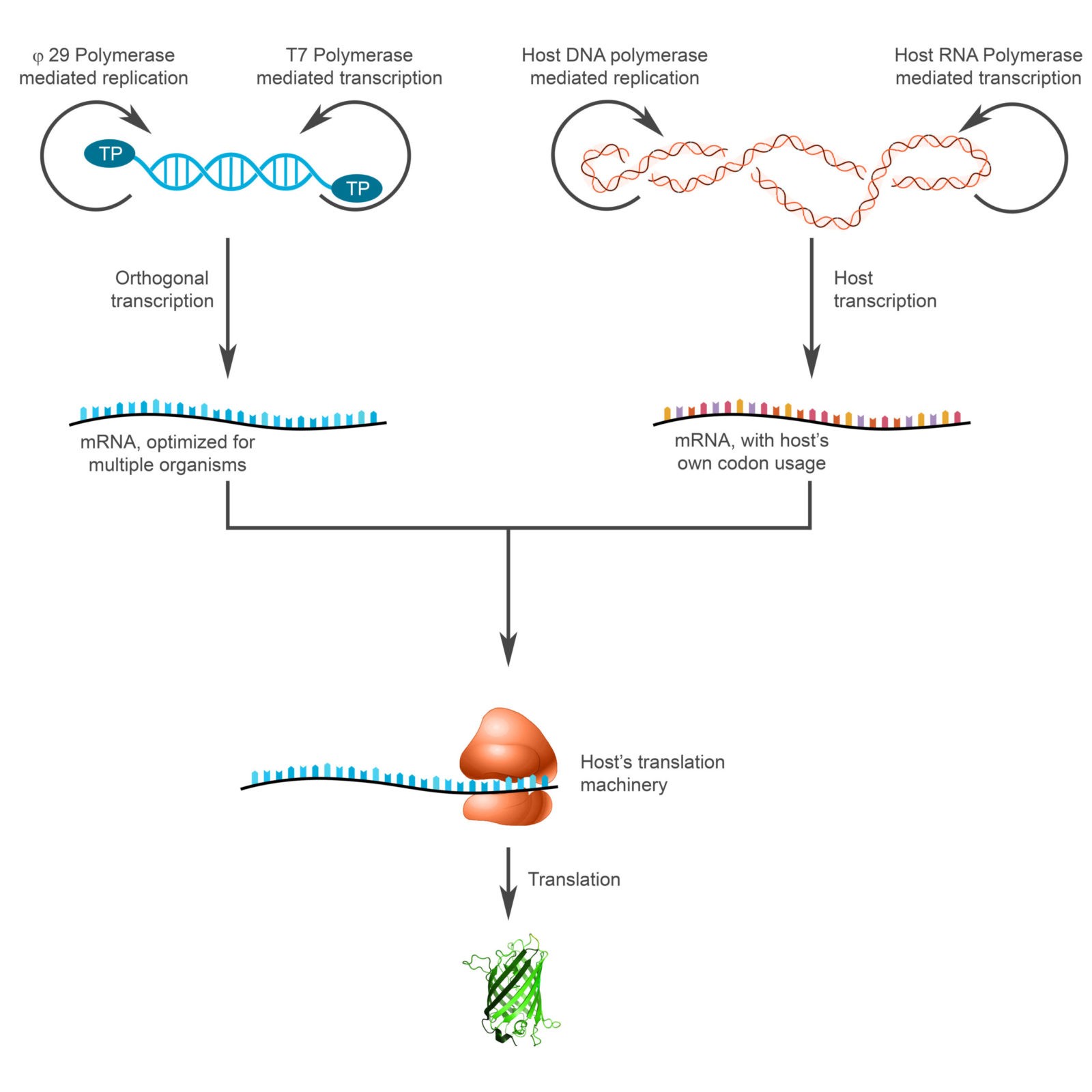
Figure 2: Orthogonal replication; our approach consists of engineering a plasmid that incorporates orthogonal replication inspired by the phi-29 bacteriophage system (linear DNA with proteins covalently bound to the 5′ termini).
The 2019 TU Delft iGEM Team
Do you want to know more about our project? Then visit our website for more information.
We are always happy to answer questions via e-mail. You can also follow us on Instagram, Facebook, and Twitter.

From top left to bottom right: Esther Hoogerwerf, Huyen My Nguyen, Weronika Hoska van Aken, Hafsa Akaouche, Sagarika Govindaraju, Renée van der Winden Marco Valentim Becker, Mu-En Lu, Osman Esen, Dennis Kenbeek, Joel Clotet i Peretó
By Joel Clotet I Peretó
Did you like this article? Then subscribe to our Newsletter and we will keep you informed about our next blog posts. Subscribe to the Eurofins Genomics Newsletter here.





