
Figure 1: Rape infected by a fungus.
The natural and modified function of the M13 phage
We, the iGEM Team Bielefeld-CeBiTec 2019, focus on developing a system to fight plant infecting fungi. We aim to create a system based on the M13 phage. The outstanding feature of the M13 phage is that it is customisable and highly specific for one eukaryotic pathogen (Figure 2). Fundamentally, M13 is a filamentous phage that infects the intestinal bacteria Escherichia coli. The DNA of a M13 phage is single stranded, and it contains an origin of replication and eleven different genes that encode the essential functions of the phage (Smeal et al., 2017). During an infection, M13 does not cause direct cell damage. Instead, it integrates its phage DNA into the infected cell, allowing replication of the phage DNA inside the infected cell. However, for our system we strip the phage of all the functions that make it an effective phage and only use its protein coat as a shuttle for our Cell Death Inducing System (CeDIS), containing Cas13a and a specific guide RNA cluster (Liu et al., 2017).
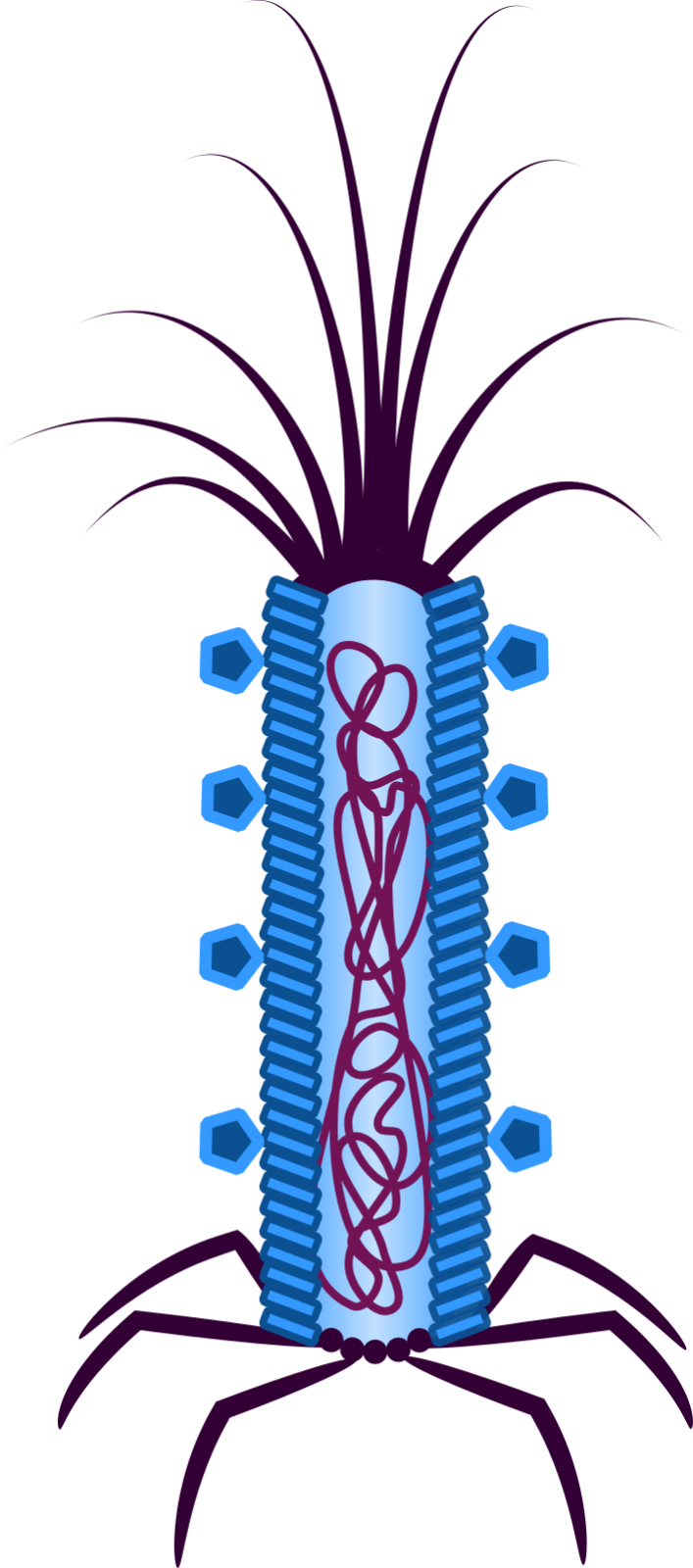
Figure 2: The phage-based platform Troygenic. Pathogen-specific ligands are located on the coat (illustrated by pentagons).
The trojan horse tactic
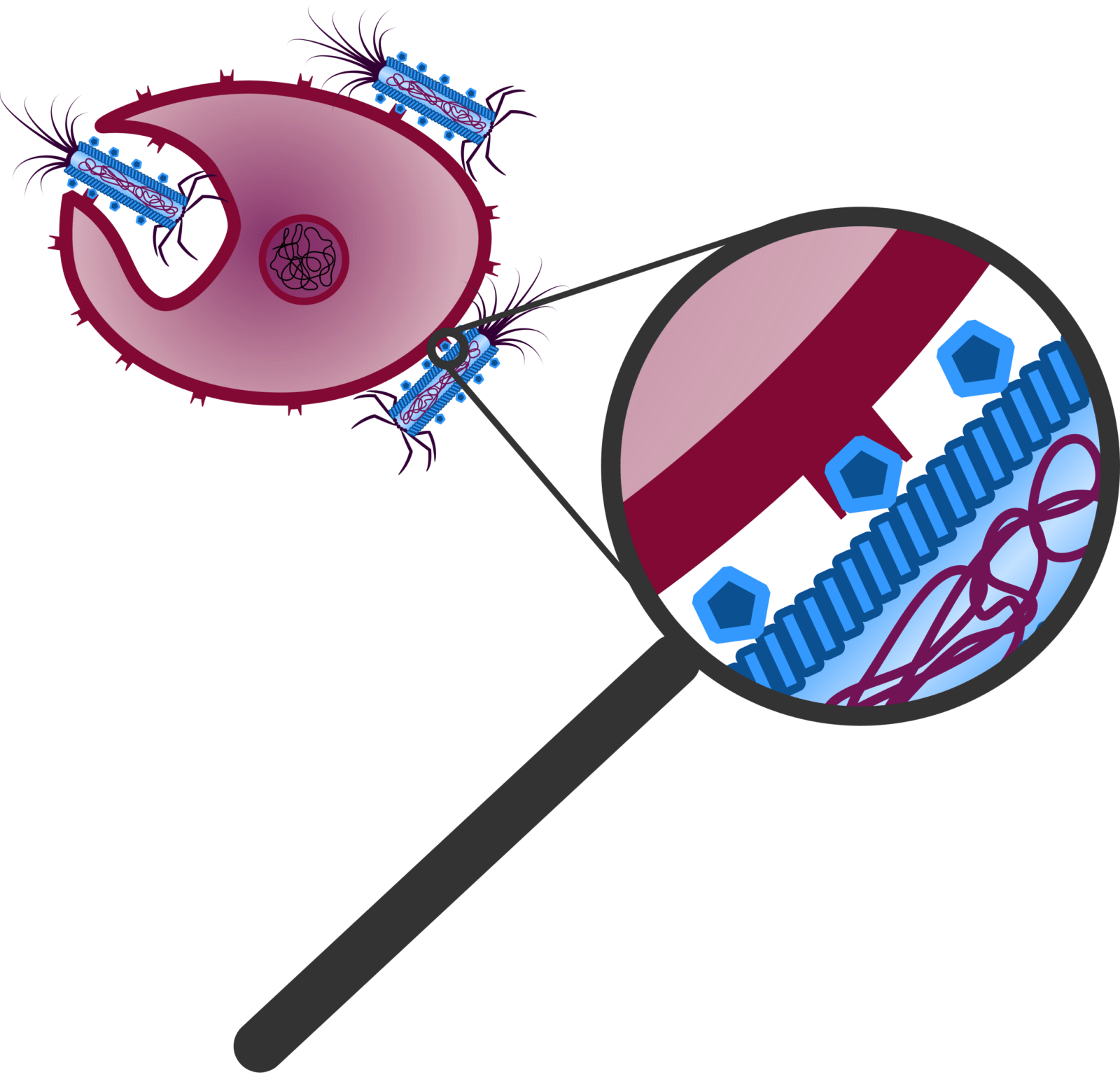
Figure 3: The ligands of the Troygenic bind to for the pathogen specific receptors and trigger endocytosis by the pathogen.
Our modified phage is called Troygenics. It has no longer the ability to infect bacteria and replicate independently. Due to its adaptable protein coat, it is specifically recognised by the pathogen of interest and taken up via endocytosis (Figure 3) (Doherty et al., 2009). CeDIS in Troygenics can also be adjusted to the pathogen of interest. In case of specific identification and binding of the pathogen’s target gene by the guide RNAs, collateral cleavage of the pathogen’s RNA is induced by Cas13a and eventually leads to cell death of the pathogen (Figure 4). With these building blocks, our Troygenics will be able to specifically identify and kill the targeted pathogen, while the host cells and other cells will remain completely unaffected.
Ultimately, our goal is to create a system that can be adjusted by minor changes to fight eukaryotic pathogens in general. This includes not just a variety of plant pathogens, but also animal and human pathogens, such as the parasitic protozoa trypanosomes that cause the sleeping sickness.
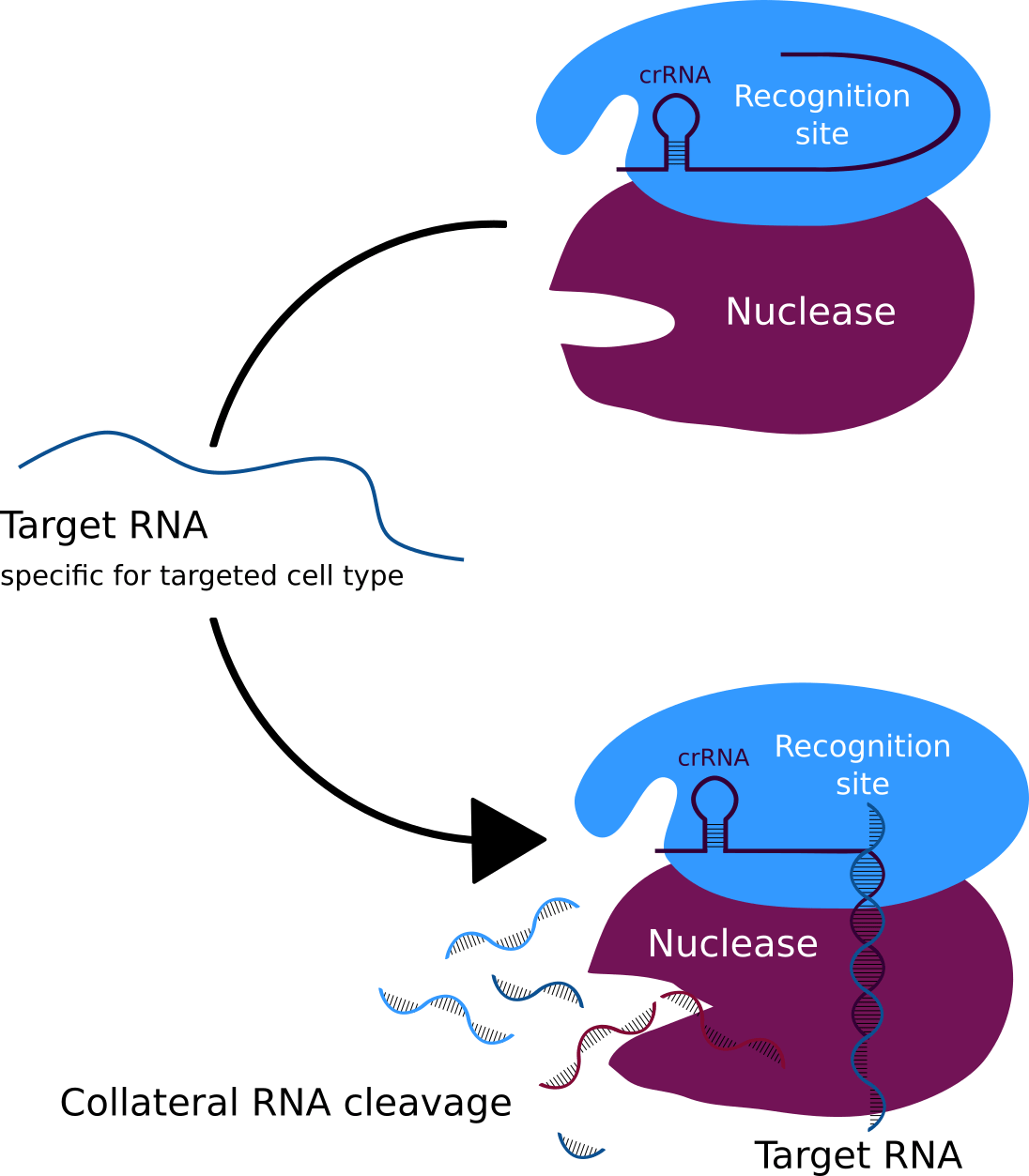
Figure 4: The CRISPR system that binds the target RNA and collateral cleavage of the pathogens RNA.
Who are we and what is iGEM?
We, the iGEM team Bielefeld-CeBiTec 2019 consists of nine highly motivated students from different faculties of Bielefeld University (Figure 5). The courses of study represented in our team are molecular biotechnology, biology and biochemistry. We are participating in this year’s international iGEM competition, the most renowned, non-commercial science competition in the field of synthetic biology that is organized by the iGEM (international Genetically Engineered Machine) foundation (Boston, USA). Students from all around the world join the competition. Last year, over 350 teams from 42 countries participated.

Figure 5: The iGEM Team Bielefeld-CeBiTec from left to right: Astrid Toebber, Niklas Fante, Isabel Conze, Johanna Opgenoorth, Katharina Wolff, Ina Schmitt, Adila Apsara, Alexander Schulze, Nefeli Chanoutsi
Our motivation
Driven by interdisciplinary, great experience in different areas of biology, as well as high levels of passion and motivation, we work hard to turn innovative ideas and methods into a successful project. Participating in iGEM allows us to further our knowledge and to discover and explore new topics. Furthermore, it gives us the chance to represent Germany as a hotspot for leading education and research on a global level. So far, the Bielefeld iGEM teams from 2010 to 2018 have achieved this with great success. Ultimately, like every team before us, we work completely independently and in addition to our regular studies on this time-consuming project that is fully planned and executed by us.
Do you fancy a great evening of education and fun?
In cooperation with the Bielefeld Natural History Museum, we have organised an exhibition in the field of synthetic biology. If you would like to learn more about the fascinating field of synthetic biology, its development and application in everyday life, come and visit us at the Bielefeld Natural History Museum. The exhibition can already be visited. We are excited to learn how you enjoyed your visit.
Do you want to know more about our project and what we are up to? Then visit our website. We are always happy to answer questions via e-mail. You can also follow us on Instagram, Facebook, and Twitter.
By Nefeli Chanoutsi and Dr Andreas Ebertz
Did you like this article? Then subscribe to our Newsletter and we will keep you informed about our next blog posts. Subscribe to the Eurofins Genomics Newsletter.





