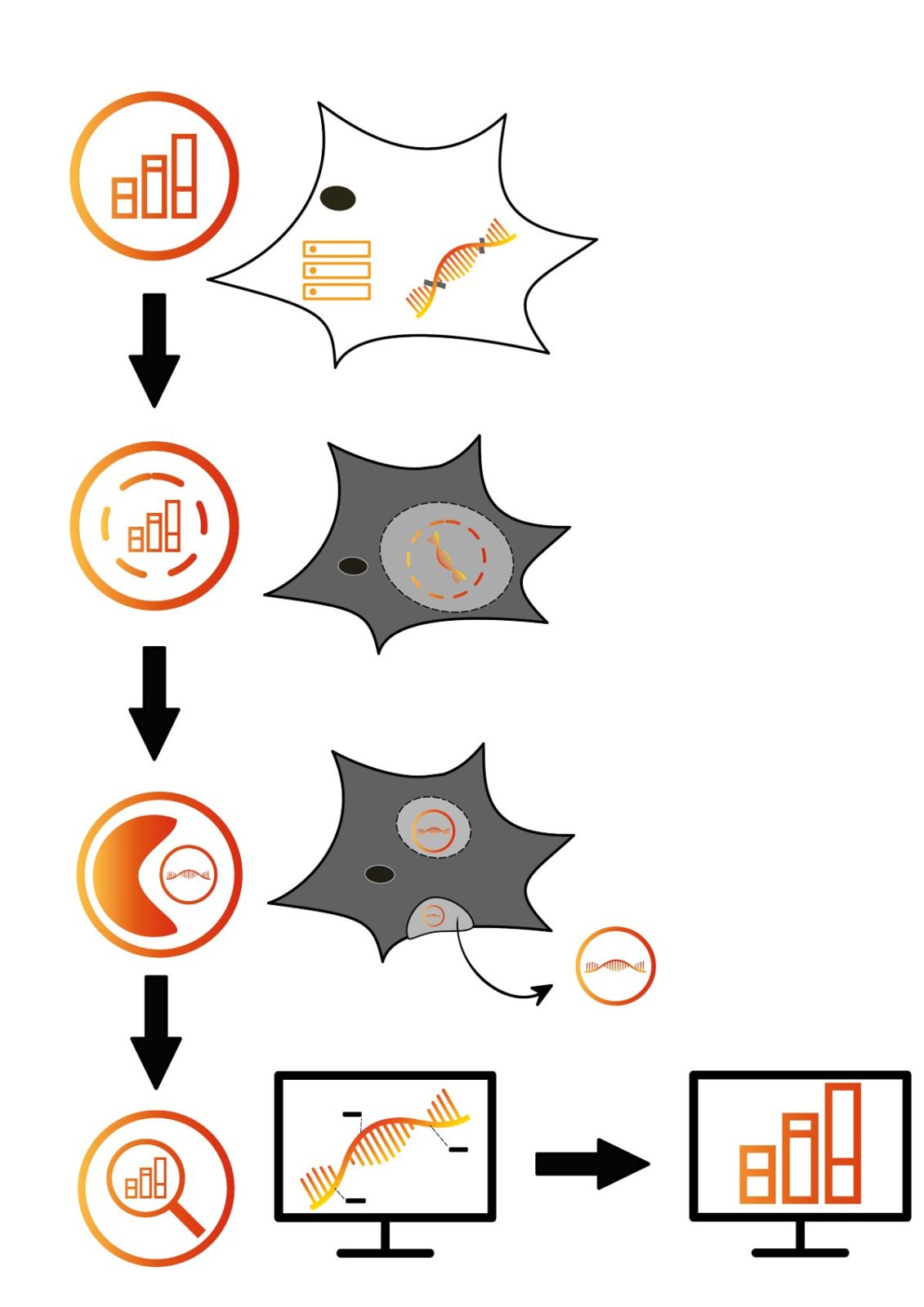
Encoding information in mRNA enables safe export from cells in virus-like particles or exosomes
In order to monitor cells minimal-invasively, we aim to manipulate them to export RNA molecules packed safely in vesicles, which can be isolated easily from fluids such as blood and then can be analysed in downstream processes.
Therefore, we encode a unique identifier in the intron of a gene of interest that is not translated and therefore does not influence the cell’s metabolism. In addition, silent mutations can be added as additional barcodes to allow single-cell resolution. The mRNA, which contains both identifier and barcodes, is directed into a protective cargo vesicle post-transcription.
Currently, we explore virus-like particles (VLPs) as a biorthogonal approach (Titeca et al. 2017), as well as exosomes. Exosomes have the advantage of not inducing immunogenicity (Kojima et al. 2018). Our VLPs consist of Gag-monomers that are structural units of the HI-virus. But other than budding, no viral functions are present in those VLPs. Thus, they are completely harmless (Titeca et al. 2017). Exosomes are part of the endogenous secretory pathway with direct cell-type dependent functions, such as intercellular communication (Kojima et al. 2018).
Those extracellular cargo vesicles are purified from the supernatant of the surrounding media via ultracentrifugation or affinity chromatography (Titeca et al. 2017). After lysis, the mRNA cargo can then be integrated into downstream analysis workflows such as RNA quantification or sequencing (Ronald et al. 2016). Thus, partial transcriptomics and monitoring of the cell state can be disconnected from invasive cell disruption while allowing in vivo reconstruction of tailored cellular processes.
Monitoring cell transplants
Apart from a broad range of applications in basic research, e.g. for cell and tissue cultures or organoids, our platform could evolve to a versatile tool for a variety of other fields, ranging from (pre-) clinical research to industrial applications, such as fermenters, all the way to regenerative medicine. We are especially enthusiastic about the latter, as cell replacement strategies are promising approaches to treat numerous diseases and increase the quality of life in affected individuals significantly.
Recently, new therapeutic techniques have emerged in the field of regenerative medicine to fight widespread diseases such as cancer and diabetes. Genetically modified T-cells such as CAR-T-cells present a valuable new immunotherapeutic treatment for several cancer types but also hold the potential to cause life-threatening side effects such as cytokine release syndrome (CRS) and anaphylaxis (Maus et al. 2013; Bonifant et al. 2016). These critical complications require precise monitoring of the CAR-T-cells to enable an immediate, life-saving treatment (Neelapu et al. 2018; Bonifant et al. 2016).
To fight diabetes, β-cell transplantation could present a long-term solution for patients. β-cells can differ in their glucose responsiveness and adaptations after β-cells islet transplantation. It is of big interest to closely monitor these different cells in the patient in order to verify their long-term functionality (Pipeleers et al. 2017).
Overall, we hope to develop a method that allows monitoring transplanted cells effectively by a simple blood sample. By choosing from the different tools that we plan to develop, researchers will be able to choose individual solutions for in vivo cell monitoring, tailored to their specific needs.
The iGEM LMU & TU Munich Team
As an international team, we communicate in the main scientific language – English. But the diversity of our team extends beyond variety in language and culture. One example is everyone’s field of study: The scientific backgrounds of most members are biology, biochemistry, and molecular biotechnology, but there are also students from the fields of bioinformatics and electrical engineering. The scientific backgrounds of our supervisors allow them to provide a different perspective that is useful to achieve our ambitious goals. With more than half of the team members pursuing their master’s degree, a great learning environment is created. Bachelor students learn from more advanced members and benefit from their pool of knowledge, whereas the younger students provide new insights and a fresh perspective.

Do you want to know more about iGEM Munich’s project? Then visit their website for more information.
The team is always happy to answer questions via e-mail. You can also follow them on Instagram (igem.munich), Facebook (iGEM LMU & TU Munich), and Twitter (iGEM_Munich).
By Johanna Wallner, Sarah Brajkovic, Joshua Hesse, Susanne Pettinger, and Prof. Gil Westmeyer
Did you like this article? Then subscribe to our Newsletter and we will keep you informed about our next blog posts. Subscribe to the Eurofins Genomics Newsletter here.