It has been established that methane is a more potent greenhouse gas than CO2 and that it plays a major role in the greenhouse effect responsible for climate change. One of the major sources for methane is livestock farming (Heilig, 1992; Reisinger and Clark, 2018). Besides methane production, livestock also requires large areas of land and high amounts of antibiotics, and produces lots of waste (Sharma et al., 2015). So, what would be an alternative to cow milk? Plant-based milk alternatives often do not achieve the goal of replacing cow’s milk because they are too different in taste.
We, the iGEM team of the Heinrich-Heine University in Düsseldorf, have put us to the task of creating cow’s milk without using any animal products. A product that is the same in taste, consistency and overall nutritiousness.
This is how we do it
We are aiming to synthesise cow’s milk by using heterologous expression of proteins, enzymes, as well as metabolic engineering, mathematical modelling and many other methods of synthetic biology. Cow’s milk is made up of lactose, lipids, proteins and nutrients, all of which are emulsified in water. As nutrients can be bought as pure chemical syntheses but the proteins and lipids from milk are not commercially available without resorting to milk isolates, we will focus on these two components.
Protein production
Milk proteins are usually divided into two categories: caseins, which are proteins that are very insoluble and occur in cheese, and whey proteins, which are mainly globular proteins that are somewhat soluble in water and get filtered out during cheese production. Caseins make up around 80% of the total protein in cow’s milk and they help with the digestion of milk by forming aggregates. Due to their high hydrophobicity, they form micelles, which help to keep calcium ions soluble and available for digestion.
Whey proteins consist of a lot of diverse proteins. We will only focus on the two most abundant proteins: ɑ-lactalbumin and β-lactoglobulin.
Caseins, on the other hand, consist of just four different proteins: ɑ-s1-, ɑ-s2-, β- and κ-casein. We will heterologously express all six of these proteins in multiple production organisms including well-known organisms like E. coli and B. subtilis, as well as in the cyanobacterium Synechocystis sp. PCC 6803 to explore the possibility of protein production in a photosynthetic organism to additionally reduce CO2 as cyanobacteria use CO2 as a carbon source.
Our main focus for yield-oriented production is on B. subtilis. This gram-positive bacterium will express our proteins fused to a peptide signal that causes the export of the protein across the cell membrane into the culture medium after which the signal peptide is cleaved off. Peptide signals for secretion are diverse and specific signals are needed for specific proteins. To get the highest possible secretion yield, we will test five different peptide signals for each of the six proteins we focus on and we will screen the culture medium for the highest concentration of these proteins. In this way we hope to find the best-suited peptide signal for each protein and get high protein yields.
For Synechocystis, we are exploring the use of a photobioreactor for a more efficient protein production per cell and the possibility for industrial large scale production in the future.
Fatty acid production
The lipids in milk are triglycerides of varying lengths, most of them range from 14-20 carbons in length, but there are also shorter lipids ranging from 4-12 carbons in length.
Long-chain fatty acids that can be converted into triglycerides are naturally produced in most microorganisms but are only secreted if they are present in abundance in the cells. To increase the yield of long-chain fatty acids in the cells, we aim to remove a bottleneck of fatty acid synthesis by overexpression of the ACC enzyme that is responsible for the reaction from Acetyl-CoA to Malonyl-CoA.
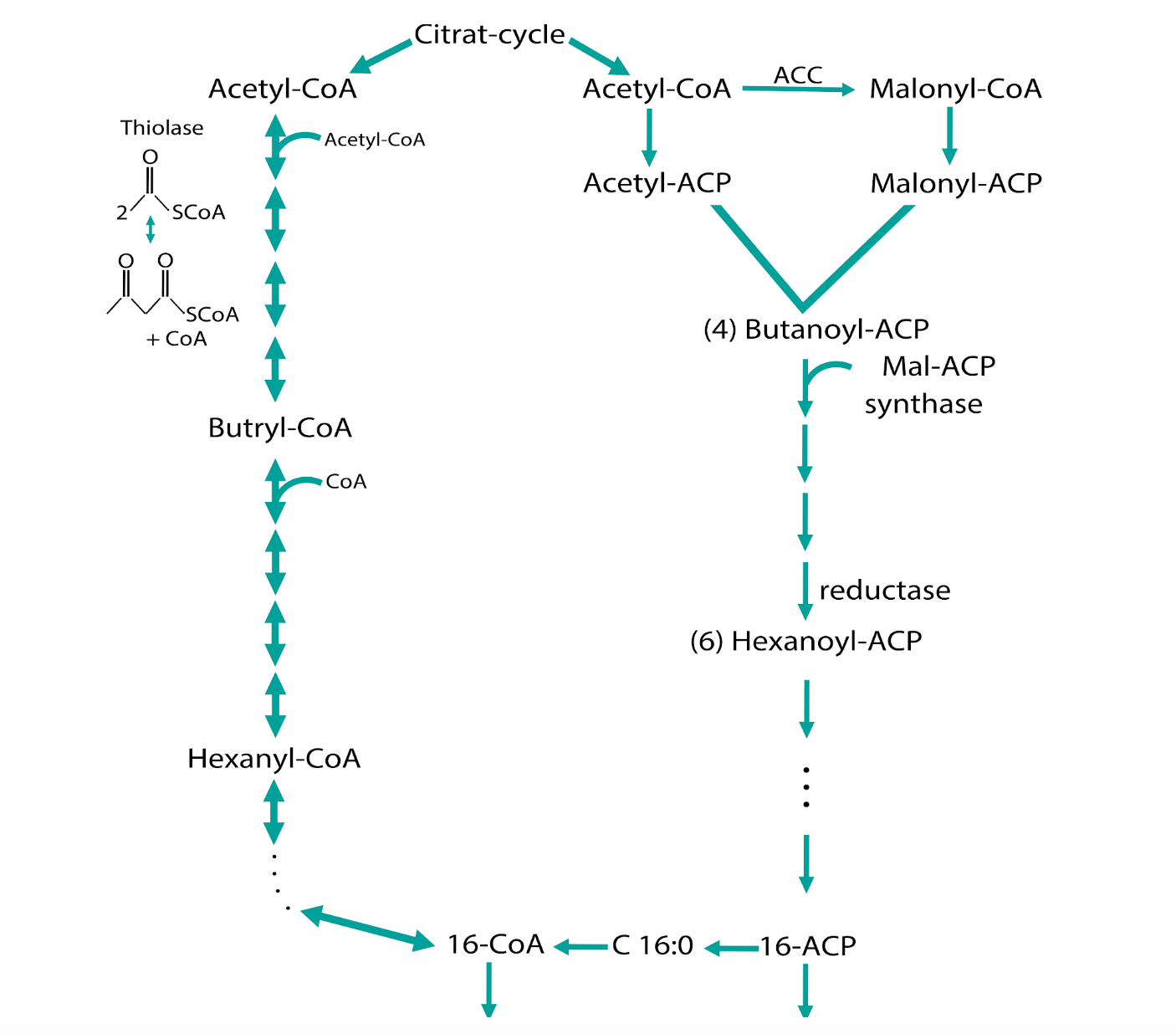
Figure 1: The synthesis and degradation of fatty acids in bacterial cells
During the basic fatty acid synthesis (Figure 1), Acetyl-CoA from the Citrate-cycle is used to produce fatty acid chains with the length of 16 or 18 carbon molecules for example (palmitic acid and stearic acid). Here, the reaction from Acetyl-CoA to Malonyl-CoA is the rate-limiting step that is regulated by the Acetyl-CoA-Carboxylase complex (ACC).
This complex consists of the 4 subunits a- and b-carboxyltransferase, one acetyl-CoA-biotin carboxyl carrier protein and one biotin carboxylase that are encoded by 4 genes: ACC A-D. We are currently overexpressing these 4 genes in E.coli DH5a and BL21DE3 to establish a proof-of-concept that the overproduction of the ACC complex is strong enough to produce fatty acids that we then can isolate from E.coli and does not interrupt the growth of the bacterium. The next step will be the overexpression of the ACC complex in the cyanobacterium Synechocystis sp. PCC 6803.
We will produce the short- and long-chain lipids separately due to their different availability in microorganisms. In order to produce short-chain fatty acids in the cells, different methods have to be employed since they do not get produced naturally in most organisms. To produce short-chain fatty acids, we will heterologously express enzymes called thioesterases that end the fatty acid synthesis at specific chain lengths.
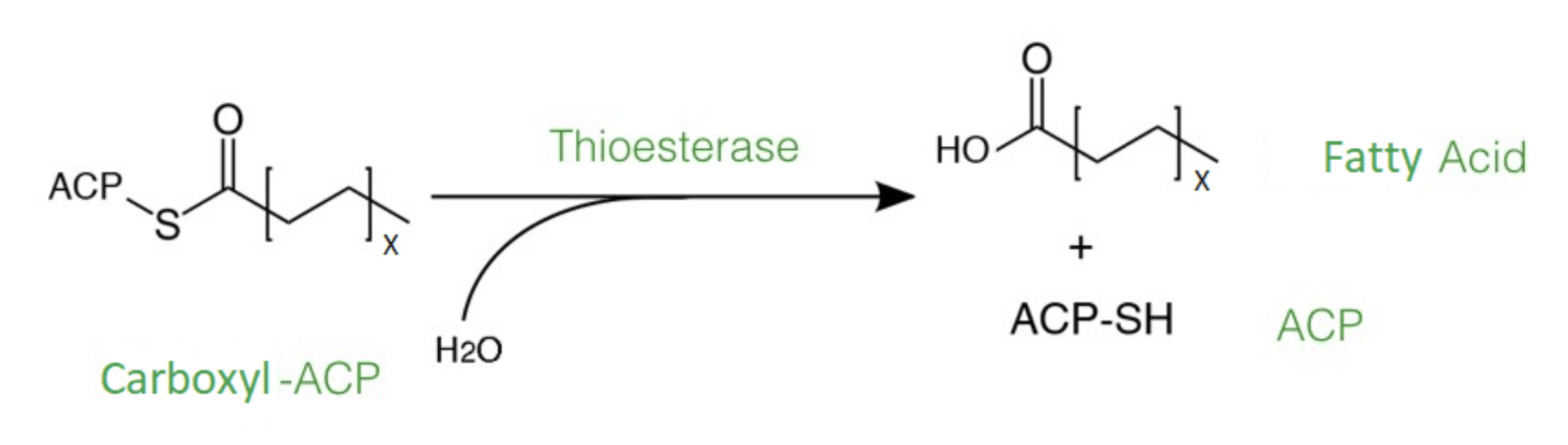
Fig. 2: The reaction catalysed by thioesterase enzymes, ending fatty acid synthesis at a specific chain length.
Fatty acids are produced and elongated by enzymes of the fatty acid synthases complex with acetyl-CoA or acetyl-acp and NADPH in the cytoplasm of the cell (Figure 2). The elongation is termined by thioesterases that hydrolyse specific acyl-CoA or acyl-ACP, which are basically activated fatty acids, to the corresponding fatty acid and coenzyme A or the acyl-carrier-protein. The thioesterases that we chose originate from different organisms, including bacteria, plants and algae. They are each specialised for ending the fatty acid elongation in an early state of the fatty acid synthesis to create the short-chain fatty acids butyric, caproic, caprylic, capric and lauric acid. The thioesterases use acyl-ACP as substrate to synthesise these fatty acids. Normally, the produced short-chain fatty acids are secreted into the medium and standard methods can be used to extract these fatty acids from the medium.
In order to obtain a higher yield, we aim to down-regulate the production of long-chain fatty acids in our bacteria. For this purpose our targets for the down-regulation are the enzymes that are responsible for the production of long-chain fatty acids, the utilisation of acyl-CoA, and the degradation of free fatty acids. As these enzymes can’t be deleted from the organism because they are part of essential metabolic pathways, we will establish an inducible down-regulation using the CRISPR-interference system with dCas9. In contrast to the CRISPR/Cas9 system, the CRISPRi is using a deficient Cas9 without nuclease activity. This leads to a binding of dCas9 to the targeted gene but without cleaving of the DNA. Due to this binding of dCas9 to the DNA, transcription of the target gene is repressed.
This method, as well as the heterologous expression of thioesterases, will then be used in the cyanobacterium Synechocystis sp. PCC 6803 to get a high yield of the desired fatty acids and to further reduce the CO2 emission, similar to our project with proteins.
For our work, we are supported with expertise by the institutes for synthetic microbiology, biochemistry and plant cell biology and biotechnology in Düsseldorf as well as the institute for molecular enzyme technology in Jülich to make our project as successful as possible.

Do you want to know more about the iGEM team Düsseldorf 2019’s project? Then visit their website for more information at https://www.igem.hhu.de/ or https://2019.igem.org/Team:Duesseldorf and sign up for their newsletter. The team is always happy to answer questions via e-mail. You can follow them onFacebook and on Twitter.
By Melanie Sbielut and Dr Andreas Ebertz
Did you like this article? Then subscribe to our Newsletter and we will keep you informed about our next blog posts. Subscribe to the Eurofins Genomics Newsletter here.





