What is our project about?
So, what’s our solution?
Determined to find the perfect organism for our endeavour, we chose the cyanobacterial strain Synechococcus elongatus UTEX 2973, due to its doubling time of under 2 hours. This could have a huge impact on utilisation, as the time consumed by many workflows is primarily dictated by the growth of the chassis. Our Synechococcus elongatus strain UTEX 2973 (Figure 1) could compete with common heterotrophic chassis like yeast, which would be a trend-setting novelty in photosynthetic research. We are dedicated to developing this strain as a chassis for the scientific community and future iGEM teams.

Figure 1: Two cultures of Synechococcus elongatus UTEX 2973 in their typical green colour.
In the third and last step, the strip will be transferred to a solution with HRP substrate. When the fusion protein dCas9-HRP has bound to the DNA of interest, it will be transferred to the third solution where a colour change of the HRP substrate will be triggered. The colour change will only occur when the sequence of interest is present.
How do we achieve that?
An important characteristic of a good chassis is that it can easily be genetically modified. This includes the construction of stable vectors to allow transient expression of the desired genes. However, the problem is that a lot of vectors do not or only scarcely replicate in cyanobacteria, especially in S. elongatus. Therefore, our goal is to free our strain of its natural plasmid pANS, allowing us to clone the origin of replication (ori) from said plasmid into our own vectors, as otherwise there could be problems in gene expression with two plasmids harbouring the same ori.
The next step would be to transform our newly built vectors into our strain. This has already been done in the more commonly used S. elongatus strain PCC 7942 due to its natural competence. So, this should not be a problem, should it?
Well, it’s not that easy in the UTEX 2973 strain. After isolation, this strain lost its natural competence due to a single point mutation, which means that the vectors would need to be transformed through other methods, such as triparental conjugation. This process would be really tiresome and way more time consuming. To tackle this issue and restore the natural competence of S. elongatus UTEX 2973, we aim to integrate a CRISPR/Cpf1 system (Figure 2) into our toolbox that enables easy genomic manipulation and, thus, gives us the tools to construct various other strains and revert the problematic point mutation. Consequently, our new strains become naturally competent again.
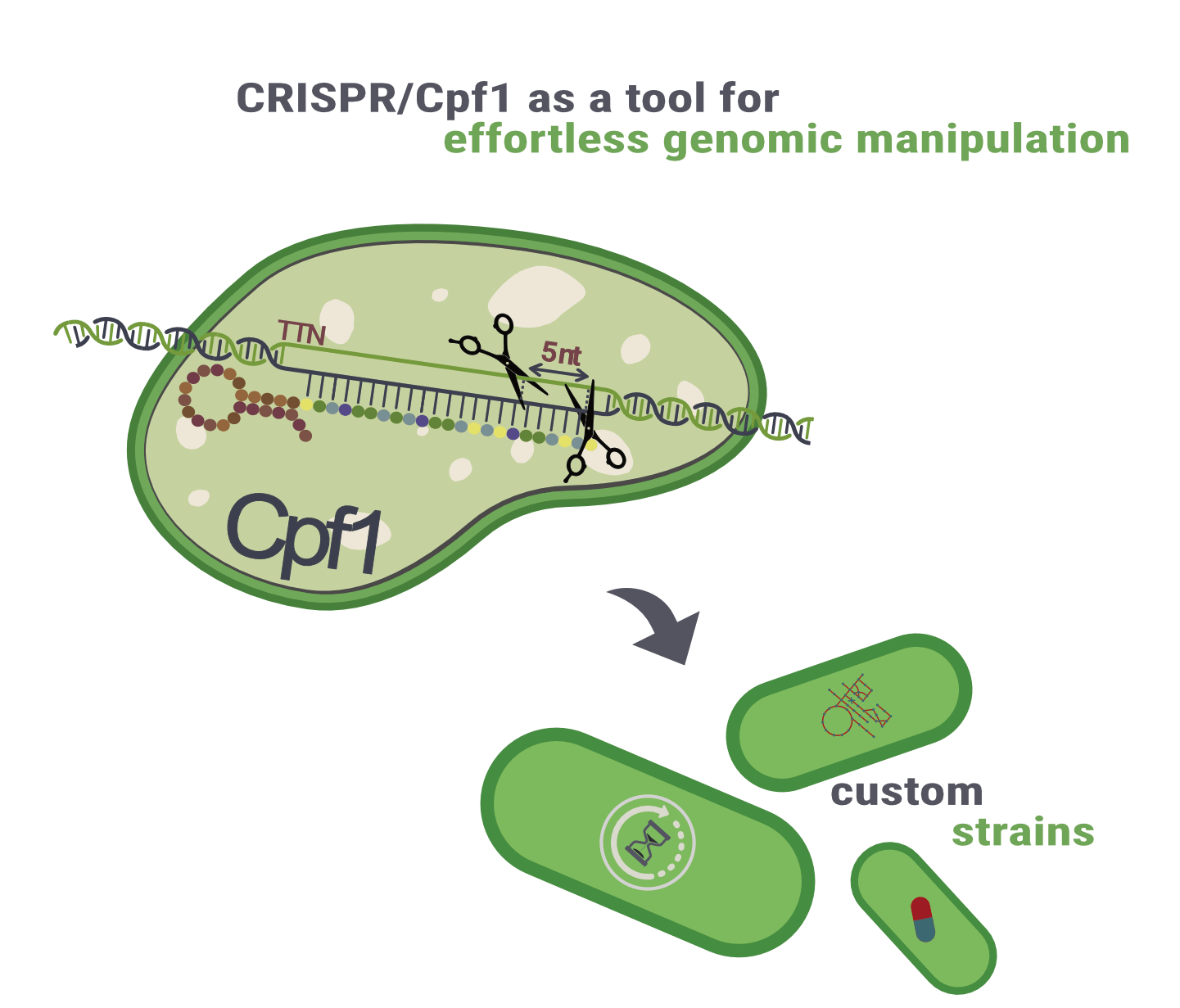
Figure 2: Derived from bacteria, CRISPR/Cpf1 is an adaptive antiviral defence mechanism that can be used to apply very precise genetic modifications – enabling us to engineer different strains.
Wait a minute… What toolbox?
Classical cloning methods are highly reliant on the restriction sites they have and that make them hardly compatible with other vectors. Changing single parts of these vectors is quite complicated and time-consuming work.
Our toolbox is a collection of vector parts based on the modular golden gate cloning method (MoClo) that facilitates the assembly of predefined parts in a modular way. This approach makes use of Type IIS restriction enzymes that allow the creation of scarless standardised overhangs – overhangs that do not include the recognition site of the enzyme anymore and thus don’t include any unnecessary base pairs (Figure 3). As every vector part is stored in a Lvl 0 vector, specific parts can be chosen freely out of the toolbox and assembled in a single tube, therefore, creating the desired plasmid with the required promoters, ribosome binding site (RBS), coding sequences (CDS), terminators, etc. in one reaction.
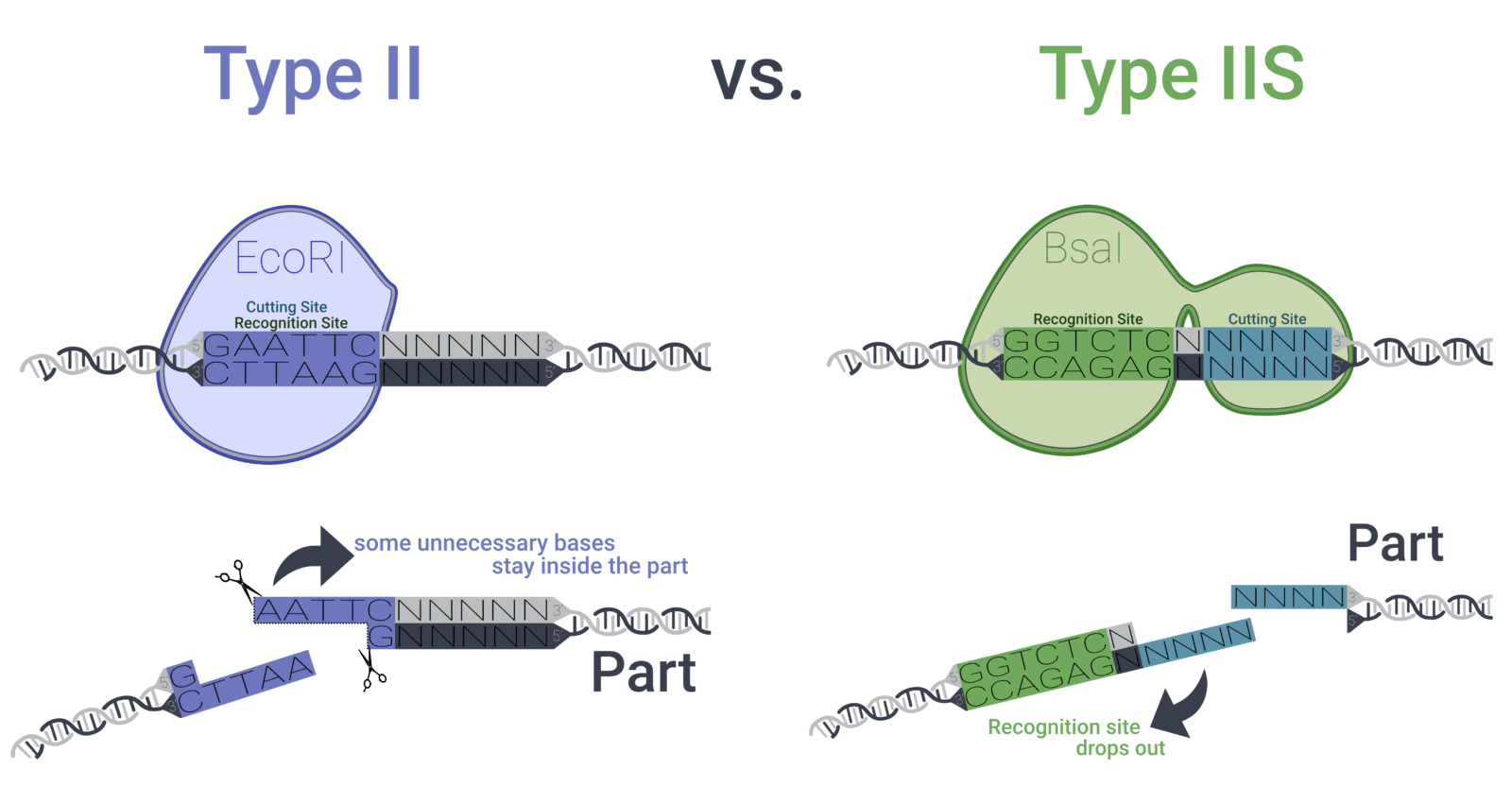
Figure 3: While the widely used type II restriction enzymes (e.g. EcoRI) cut inside their recognition sequence, leaving behind some base pairs inside the desired genetic part, type IIS restriction enzymes (e.g. BsaI) cut outside of their recognition sequence – an ideal trait for the creation of genetic parts that solely consist of the desired base pairs.
The fact that the overhangs are predefined and standardised makes it a lot easier for different iGEM teams and labs to exchange vector parts, allowing a rapidly growing collection of these parts for everyone to use. This year, we will characterise old parts and add new ones to the existing Marburg Collection for the work in S. elongatus. These new parts include some that enable the construction of whole operons using special connectors – a novelty in this modular cloning method. These connectors are small genetic fragments with overhangs that skip certain parts, such as the terminator and the promotor, allowing to go directly from one CDS to the next RBS in this modular toolkit.
To showcase the advantages of utilizing our chassis for an accelerated design-build-test cycle, we aim to create a workflow as a proof of concept for rapid testing of genetic constructs and enzymes by screening different farnesene-synthases that produce alpha-farnesene, a component of bio jet fuel.
What other ground-breaking projects outside the wet lab are we tackling?
Cloning plays a central role in synthetic biology, but it is a time intensive step in every project. There are already possibilities to automate these processes, but most of them are pretty expensive. We want to contribute to an inexpensive alternative to cloning. Last year, the iGEM Team Marburg won the Opentrons OT-2 in a request for proposals – a pipetting robot that was designed as a liquid handler, but thanks to its completely open source feature, we can customise it to fit our necessities.
Our plan is to automate the colony picking part of cloning with this robot. Here, a Raspberry Pi with an additional camera and an artificial neural network trained for colony detection will guide the Opentrons in picking the colonies that we want. To generate a consistent data set with similar colonies, a light table module for agar plates will be indispensable. This will not only ensure that the Opentrons can pick colonies independently of day and night cycles, it also enables green white selection via GFP by integrating UV-light into the Opentrons table (Figure 4). The same setup can be used for plating, and by adding an orbital shaker module even plasmid purification will be possible.
Of course, all scripts and blueprints for these modules will be freely available for the scientific community and give other teams that already own one of these robots the possibility to expand its abilities!
We would love to see other people pick up our work and continue to chase our vision of providing a full cloning processes from ordered oligos to a finished construct!

Figure 4: Inside the Opentrons OT-2: behind the pipetting arm we mounted a Raspberry Pi and a camera, so that the robot can detect colonies on agar plates, that can be mounted onto the light table on the left side – this ensures perfect illumination without unwanted shadows and reflections.
Sooo, is iGEM just lots of work?
Basically yes, but also no.
Of course, in a project like this there is lots of stuff to do and we are working on it 24/7 (that’s at least what it feels like), but it is more than just work. It is simply awesome to be involved with so many different people from all kinds of different fields who are interested in the same project! Getting to know various views and perspectives on the same issues by mathematicians, chemists, biologists, physicists etc., who don’t always think alike, can be a huge help to broaden one’s view.
Furthermore, we get the chance to meet interesting people that can help us with our research and introduce us to up-to-date topics and newly generated results. That really helps us to dive into this awesome world of science!
Apart from all the (dry) lab work, there is also other stuff going on. Together, we visit fairs and meet-ups, join scientific events and talks or engage the general public (Figure 5), trying to inform other people about what is going on in these awesome biological fields – we try to make them understand that they don’t always have to be scared of all those little microbes!

Figure 5: On this year’s “Hessentag”, a state fair in our region, we carried out small experiments and quiz games about synthetic biology, in order to reach out to the general public and bring them closer to the microscopic life around us.
Who is the iGEM Team “Syntex” from Marburg 2019?
We are 18 students of the Philipps University Marburg, Justus-Liebig University Gießen and the University of Applied Sciences (THM) Gießen from interdisciplinary fields of Chemistry, Biology, Biotechnology, Bioinformatics, Physics, Computer Science and Pharmacy. Our goal is to provide the world with an organism that accelerates research on phototrophs and sustainable production of valuable compounds, paving the way for a greener future.
What is iGEM?
iGEM (international Genetically Engineered Machine) is a competition in synthetic biology that was founded in 2003 by the renowned Massachusetts Institute of Technology. At that time there were only five teams from Boston itself, but today more than 300 teams from more than 40 countries are competing!
In this competition, young scientists get the possibility of pushing the boundaries of synthetic biology and technology, developing creative solutions for the world’s most discussed problems. Such a project holds the possibility of looking beyond the horizon of what the normal student life has to offer: working on an own project for a year, gives deep insights into how research actually works and it is an interesting way for students to get more experience in the lab and next to it, as it also includes tasks like designing an own website, managing finances and openly discussing important biological topics with the public.
At the end of this exciting year the teams gather in Boston at the Giant Jamboree to present their projects in front of thousands of enthusiastic scientists.

Do you want to know more about project of the iGEM Team “Syntex” from Marburg 2019? The team is always happy to answer questions via e-mail. You can follow them on Facebook (@IGEMMarburg2019), Twitter (@iGEMmarburg2019) and Instagram (@igem.marburg.2019) to stay in touch about their last sprint to the competition in October.
By Jonas Freudigmann, Hinrik Plaggenborg and Dr Andreas Ebertz
Did you like this article? Then subscribe to our Newsletter and we will keep you informed about our next blog posts. Subscribe to the Eurofins Genomics Newsletter here.






One Comment