We, the iGEM-Team of TU Dresden team, aim to provide a fast and easy tool for detecting any nucleic acid sequence of interest from microbial samples and human cells, so that our method can later be applied in the lab to genotype E.coli and other microbial model organisms, as well as for detecting genetic mutations/insertions in humans. By combining a novel DNA extraction method with a newly designed fusion protein, it will be possible to obtain a visual colour readout within minutes that will indicate the presence or absence of the sequence of interest. This method will be designed to be applicable in the field, which means it will be cheap, fast and not require advanced technologies. Ideally it will not even require electricity. By doing so, we will be able to open a new scope of diagnostic application to a broad scientific audience and even the general public.
This is how we will accomplish it
In the first step, DNA will be extracted directly from the raw cell lysate by immobilising it on a cellulose paper (based on a method developed by Zou et al. 2017).
After a short washing step (Figure 1) to remove proteins and membrane particles from the cell lysate, highly pure DNA with a concentration of around 40 ng/µl will be left on the paper (OD260/280 ~ 1.8).
In the second step, the cellulose paper is dipped into a solution that contains dCas9 linked to horseradish peroxidase (dCas9-HRP). The dCas9 will recognise and bind to the specific DNA sequence that is determined by its guideRNA.
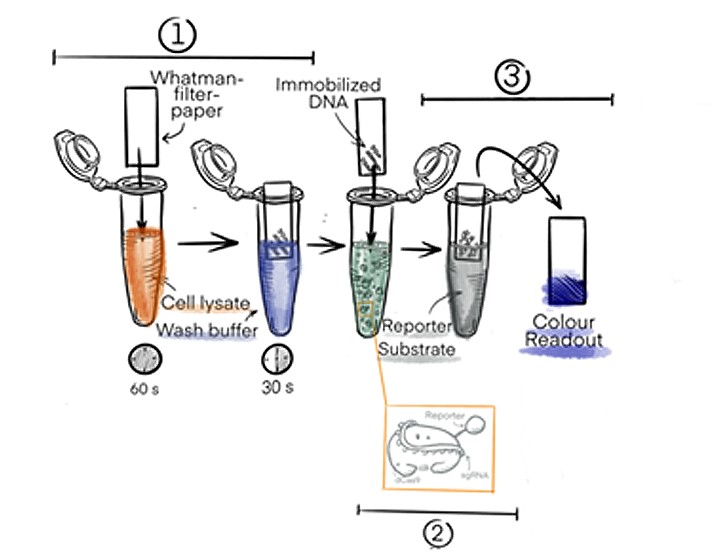
Figure 1: Basic procedure of the DipGene gene detection process. The DNA from the sample is bound to a Whatman filter paper and the presence of a specific sequence of interest is indicated by a color readout visible to the naked eye.
In the third and last step, the strip will be transferred to a solution with HRP substrate. When the fusion protein dCas9-HRP has bound to the DNA of interest, it will be transferred to the third solution where a colour change of the HRP substrate will be triggered. The colour change will only occur when the sequence of interest is present.
The Biobrick Idea – the Heart of iGEM
The heart of the iGEM-competition is a collection of so-called Biobricks, which are genes with biological functions that are flanked by a certain assembly of restriction enzymes. These make it possible to assemble the Biobricks to obtain new functions. These Biobricks can be promoters, proteins, enhancers, protein-tags, protease-cleavage sites and many more. The traditionally used prefix and suffix flanking the genes of interest was: EcoRI, NotI, XbaI upstream and Spe, NotI and PstI downstream. This was called the RFC10 standard. This standard, however, does not support translational fusions.
An iGEM-Team from Freiburg, 2008, therefore designed a new prefix and suffix that include two additional restriction enzymes, NgoMIV and AgeI. These make it possible to fuse two proteins without losing the reading frame for the downstream protein, which is normally the case, due to the scar in-between the Biobricks.
The use of NgoMIV leaves an overhang of six nucleotides that is compatible with AgeI. This overhang encodes for exactly two amino acids (iGEM Foundation, 2019a).
How the new RCF25 standard will be used to assemble our fusion-protein is depicted in a simplified version in Figure 2. The genes of interest will be amplified from the plasmids by PCR, with the primers carrying the prefix und suffix sequences. The gene fragments will be digested with the respective enzymes and ligated together in the required order. The final construct will be much more complicated than depicted in Figure 2, since additional sequences for purification of the final protein and a linker between the proteins, as well as an inducible promoter will be needed.
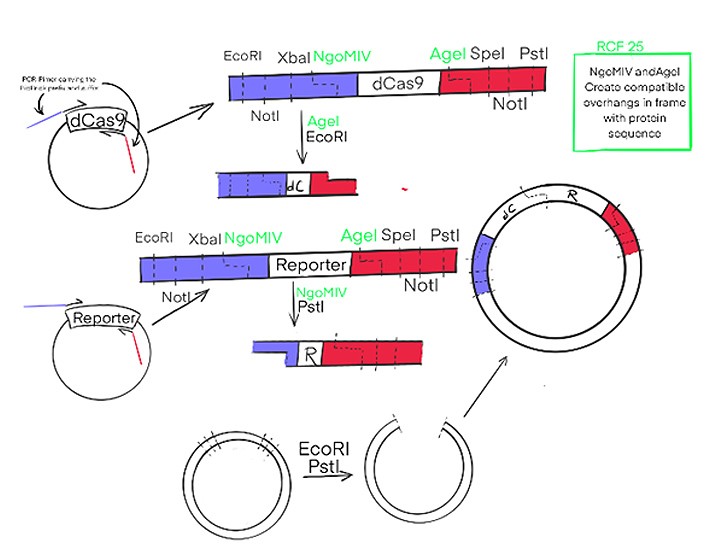

Figure 2: Cas9 and the Reporter, here HRP, will be amplified in PCR with primers carrying the sequence of the RCF25 standard. This way DNA-fragments will be obtained with the genes of interest and four different restriction enzymes upstream and downstream; these can be used to assemble the genes in any order of interest to form fusion proteins. The RCF25 standard is a new Biobrick standard that allows for translational fusion. While the black ones are from the original RCF10 standard. The Insert emphasises the advantage of the new.standard.
Every year, iGEM teams receive a distribution kit that contains a selection of the best Biobricks that all the previous teams have created (iGEM Foundation, 2019b). These are used the realise the ideas of the current teams, which will add new Biobricks to the collection.
For the future development of our diagnostic technique, the basic idea of the Biobrick assembly comes in handy. As this will enable us to use the dCas9-construct, flanked by the Biobrick prefix and suffix, and attach different reporter enzymes like alkaline phosphatase or glucose oxidase. The genetic parts are designed in a way that makes their recombination into different constructs convenient, easy and fast (NEB, 2019). This will make it possible to get a readout in different colours. With each reporter catalysing a colour reaction for a different substrate, one could test for several different disease genes at once. For example, for the disposition for breast cancer and Alzheimer’s or the integration of genes of the HI-Virus in the human genome.
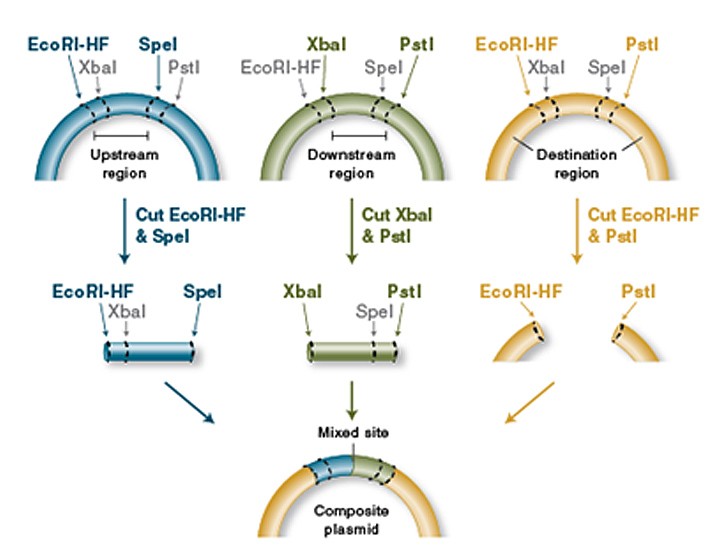
Figure 3: Basic principle oft he Biobrick assembly (here depicted in the RCF10 standard) (NEB, 2019).
For laboratory applications, it could additionally be of interest to fuse reporters to a variety of Cas-proteins that then can be used to target either RNA or DNA as needed. This opens the possibility for several applications in the laboratory such as targeting mRNA to study the human or microbial transcriptome to address changed gene expression levels in disease states. The method can also be used to genotype model organism, screen bacteria for successful integration of a transgenic construct without having to wait for one and a half hours of PCR followed by a gel run. Due to the easy adaptability of this process, to target any sequence of interest, specified by the guideRNA, the applications are basically unlimited.
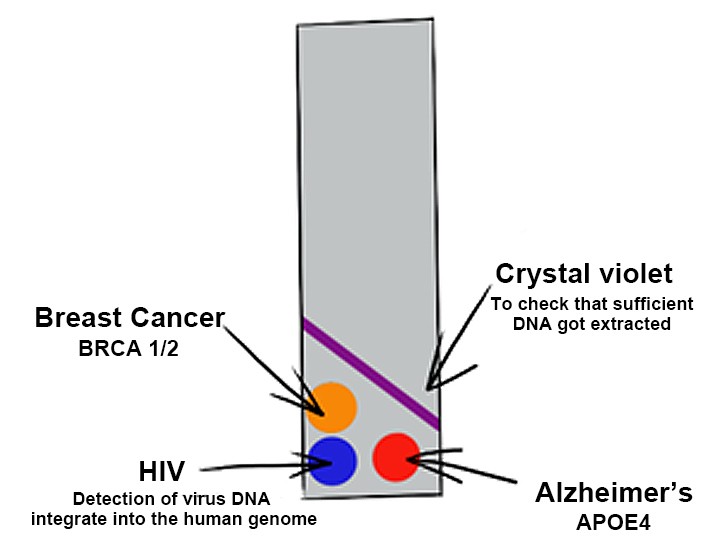
Figure 4: A potential future extension of the project, in which one single test can give readouts in different colors for several disease genes.
Our Team
This project is part of this year’s iGEM competition from the team of TU Dresden.

From left to right – Upper row: Behbood Moeini, Victoria Sarangova, Sophie-Luise Heidig, Nikitha Vavilthota, Juan Sebastian Eguiguren and Anastasia Kurzyukova – Lower row: Mara Müller, Pedro Guillem, Paula Santos Otte, Arnau Pérez Roig
Do you want to know more about project of the iGEM Team Dresden 2019? Then visit their website for more information.
The team is always happy to answer questions via e-mail. You can follow them on Instagram (igem_tudy) and Twitter (@igemTUDY).
By Mara Müller and Dr Andreas Ebertz
Did you like this article? Then subscribe to our Newsletter and we will keep you informed about our next blog posts. Subscribe to the Eurofins Genomics Newsletter here.






One Comment