A Ray of Hope
However, for some inherited diseases it is not imperative to modify all cells to cure or reduce the symptoms of the disease. CRISPR-mediated gene editing of just enough cells to make sufficient lactase could make it possible for lactose intolerant people to enjoy some proper ice cream with milk instead of this vegan and sugar-free rubbish.
In mature organisms, viral vectors such as an adenoviral vector are used to deliver the necessary genes for gene editing. This kind of sounds like one of the Bourne movies, where Jeremy Renner infects himself with a genetically modified virus to reteain his superhuman powers. In reality, it does not work that great though. In a recent publication, this approach was chosen to selectively replace the mutated Rho gene with the wild-type gene to ameliorate, not cure, the progression of retinitis pigmentosa in mice (Tsai et al., 2018).
In order to generate a systemic modification, cells that produce sperm or eggs, or are spem or egg themselves (germline cells) need to be targeted. The modification will then not only apply to the whole body but will also be passed on to the next generation.
CRISPR-mediated gene editing can not be used as a therapeutic in mature organisms. The current state of the art of CRISPR technologies faces the issue of delivering all the necessary components for gene editing to all the cells (systemic gene editing).
The biggest issue is delivery
Accuracy is of paramount importance in gene editing. With CRISPR, accuracy depends on the high target specificity of the utilised gRNA. A low specificity usually leads to alterations of unintended genome sites in addition to the gene of interest. As the saying goes: “Garbage in, garbage out”.
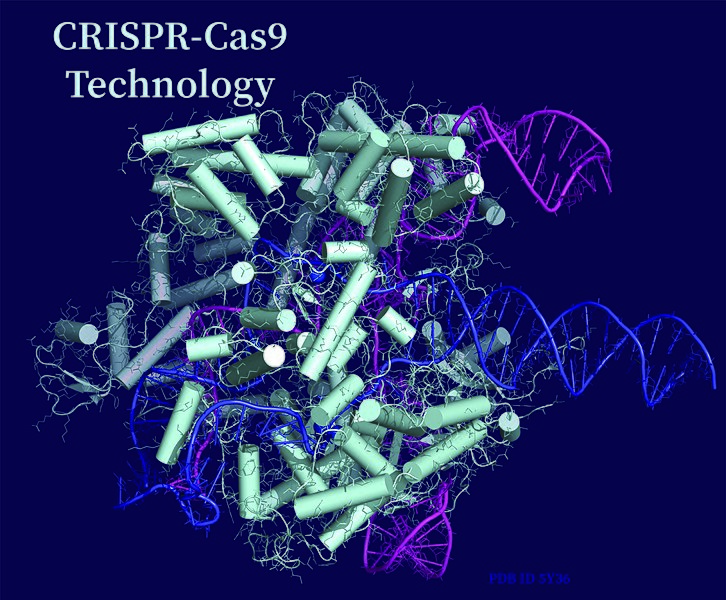
Accuracy – the Crux of the Matter
Many laboratories still use in vitro transcribed (IVT) gRNAs, despite the fact that IVT-made gRNA varies in pureness. It often contains unintended fragments and transcripts that lead to off-target effects and high variability between experiments. Synthetic sgRNAs, on the other hand, produce much more consistent results due to their optimised synthetic production.
Eurofins Genomics provides the high-quality synthetic single guide RNA (sgRNA) that guides your CRISPR experiment to success. These synthetic sgRNAs generate much more efficient (indel frequencies of up to 97%), and consistent gene edits compared to other types of gRNAs. Single guide RNA (sgRNA) has become the most popular format with researchers.
Do you need chemically modified synthetic sgRNAs to increase stability and reduce activation of immune responses (Wienert et al., 2018)? We’ve got you covered!
Check out our services here.
Confirmation of the CRISPR-mediated genome alterations and editing efficiency are of critical importance for a study. Eurofins Genomics also provides you with the right end-to-end services: Custom DNA Sequencing.
But wait, there is more!
Be aware of this befor you design your gRNA
CRISPR-mediated gene editing with Cas9 fails about 15% of the time. This is caused by a failure of Cas9 to detach from the DNA after introducing the double strand break (DSB). Subsequently, the cell’s DNA repair machinery is blocked from accessing the DSB. However, translocating RNA polymerase can knock Cas9 off the DNA and allow for the repair of the DSB in bacteria and mammalian cells. This only works when the gRNA binds to the strand of the double stranded DNA (dsDNA) that is utilised as template by the RNA polymerase. Due to more effective use of Cas9, less of the enzyme could be needed, and the interaction time of Cas9 and DNA is reduced, which could decrease the chance of adverse effects (Clarke et al., 2018).
In mammalian cell lines and mice, the CRISPR-mediated gene edit efficency was significantly enhanced by co-injection of Scr7, an inhibitor of DNA ligase IV that is a key enzyme in non-homologous end joining (NHEJ), and, therefore, promoting homology-directed repair (HDR). The utilisation of Scr7 increased the the insertion of short and long DNA fragments in the target loci by up to 19-fold (Maruyama et al., 2015).
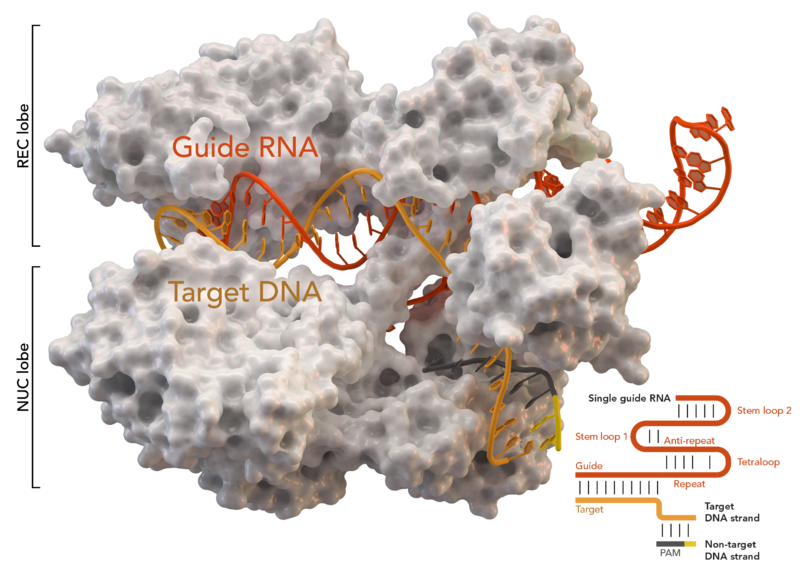
Have you also thought about changing Cas enzymes? Even though Cas9 is the gold standard CRISPR enzyme, it is known for its off-target DNA binding. The first seven to eight target nucleotides are the decisive factor for sufficient Cas9 DNA binding. After that, Cas9 seems to pay much less attention to mismatches. This of course increases off-target genome edits. Cas9-nickase (Cas9n) requires two gRNAs in combination to introduce a DSB, which also significantly increases specificity (Jinek et al., 2012). This is the preferred approach for CRISPR-mediated gene editing.
Conclusion
As of now, CRISPR-mediated gene editing is not the magic solution that will solve all of the world’s problems. But saying it is only a lab technique for more efficient testing of hypotheses would do it injustice.
Nevertheless, there are exciting times ahead in the field of genomics, and CRISPR technology could really change the world some day. It definitely has the potential! To make this dream come true, we provide your research the guide (RNA).
If you are interested in cooperating with us for a publically funded project, get in touch with our Research and Development department: www.eurofinsgenomics.eu/collaborations.
Did you like this article? Then subscribe to our Newsletter and we will keep you informed about our next blog posts. Subscribe here.